Protecting the proteome: Eukaryotic cotranslational quality control pathways
- PMID: 24535822
- PMCID: PMC3926952
- DOI: 10.1083/jcb.201311103
Protecting the proteome: Eukaryotic cotranslational quality control pathways
Abstract
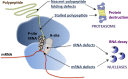
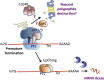
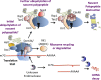
The correct decoding of messenger RNAs (mRNAs) into proteins is an essential cellular task. The translational process is monitored by several quality control (QC) mechanisms that recognize defective translation complexes in which ribosomes are stalled on substrate mRNAs. Stalled translation complexes occur when defects in the mRNA template, the translation machinery, or the nascent polypeptide arrest the ribosome during translation elongation or termination. These QC events promote the disassembly of the stalled translation complex and the recycling and/or degradation of the individual mRNA, ribosomal, and/or nascent polypeptide components, thereby clearing the cell of improper translation products and defective components of the translation machinery.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

