CDK phosphorylation of SLD-2 is required for replication initiation and germline development in C. elegans
- PMID: 24535824
- PMCID: PMC3926958
- DOI: 10.1083/jcb.201310083
CDK phosphorylation of SLD-2 is required for replication initiation and germline development in C. elegans
Erratum in
- J Cell Biol. 2014 Mar 17;204(6):1075
Abstract
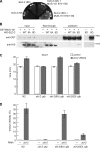
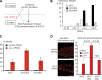
Cyclin-dependent kinase (CDK) plays a vital role in proliferation control across eukaryotes. Despite this, how CDK mediates cell cycle and developmental transitions in metazoa is poorly understood. In this paper, we identify orthologues of Sld2, a CDK target that is important for DNA replication in yeast, and characterize SLD-2 in the nematode worm Caenorhabditis elegans. We demonstrate that SLD-2 is required for replication initiation and the nuclear retention of a critical component of the replicative helicase CDC-45 in embryos. SLD-2 is a CDK target in vivo, and phosphorylation regulates the interaction with another replication factor, MUS-101. By mutation of the CDK sites in sld-2, we show that CDK phosphorylation of SLD-2 is essential in C. elegans. Finally, using a phosphomimicking sld-2 mutant, we demonstrate that timely CDK phosphorylation of SLD-2 is an important control mechanism to allow normal proliferation in the germline. These results determine an essential function of CDK in metazoa and identify a developmental role for regulated SLD-2 phosphorylation.
Figures





References
-
- Abe T., Yoshimura A., Hosono Y., Tada S., Seki M., Enomoto T. 2011. The N-terminal region of RECQL4 lacking the helicase domain is both essential and sufficient for the viability of vertebrate cells. Role of the N-terminal region of RECQL4 in cells. Biochim. Biophys. Acta. 1813:473–479 10.1016/j.bbamcr.2011.01.001 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

