Hairpin DNA sequences bound strongly by bleomycin exhibit enhanced double-strand cleavage
- PMID: 24548300
- PMCID: PMC3988684
- DOI: 10.1021/ja500414a
Hairpin DNA sequences bound strongly by bleomycin exhibit enhanced double-strand cleavage
Abstract
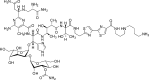
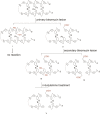
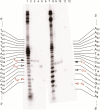
Clinically used bleomycin A5 has been employed in a study of double-strand cleavage of a library of 10 hairpin DNAs originally selected on the basis of their strong binding to bleomycin. Each of the DNAs underwent double-strand cleavage at more than one site, and all of the cleavage sites were within, or in close proximity to, an eight-base-pair region of the duplex that had been randomized to create the original library. A total of 31 double-strand cleavage sites were identified on the 10 DNAs, and 14 of these sites were found to represent coupled cleavage sites, that is, events in which one of the two strands was always cleaved first, followed by the associated site on the opposite strand. Most of these coupled sites underwent cleavage by a mechanism described previously by the Povirk laboratory and afforded cleavage patterns entirely analogous to those reported. However, at least one coupled cleavage event was noted that did not conform to the pattern of those described previously. More surprisingly, 17 double-strand cleavages were found not to result from coupled double-strand cleavage, and we posit that these cleavages resulted from a new mechanism not previously described. Enhanced double-strand cleavages at these sites appear to be a consequence of the dynamic nature of the interaction of Fe·BLM A5 with the strongly bound hairpin DNAs.
Figures






References
-
- Sausville E. A.; Peisach J.; Horwitz S. B. Biochemistry 1978, 17, 2740. - PubMed
-
- Sausville E. A.; Stein R. W.; Peisach J.; Horwitz S. B. Biochemistry 1978, 17, 2746. - PubMed
-
- Hecht S. M. Acc. Chem. Res. 1986, 19, 383.
-
- Burger R. M. Chem. Rev. 1998, 98, 1153. - PubMed
-
- Povirk L. F. In Molecular Aspects of Anti-Cancer Drug Action; Neidle S., Waring M., Eds.; Macmillan: London, 1983; Vol. 3.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

