Hepatic Hedgehog signaling contributes to the regulation of IGF1 and IGFBP1 serum levels
- PMID: 24548465
- PMCID: PMC3946028
- DOI: 10.1186/1478-811X-12-11
Hepatic Hedgehog signaling contributes to the regulation of IGF1 and IGFBP1 serum levels
Abstract
Background: Hedgehog signaling plays an important role in embryonic development, organogenesis and cancer. In the adult liver, Hedgehog signaling in non-parenchymal cells has been found to play a role in certain disease states such as fibrosis and cirrhosis. However, whether the Hedgehog pathway is active in mature healthy hepatocytes and is of significance to liver function are controversial.
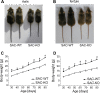
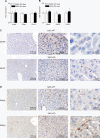
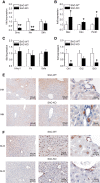
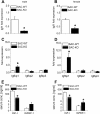
Findings: Two types of mice with distinct conditional hepatic deletion of the Smoothened gene, an essential co-receptor protein of the Hedgehog pathway, were generated for investigating the role of Hedgehog signaling in mature hepatocytes. The knockout animals (KO) were inconspicuous and healthy with no changes in serum transaminases, but showed a slower weight gain. The liver was smaller, but presented a normal architecture and cellular composition. By quantitative RT-PCR the downregulation of the expression of Indian hedgehog (Ihh) and the Gli3 transcription factor could be demonstrated in healthy mature hepatocytes from these mice, whereas Patched1 was upregulated. Strong alterations in gene expression were also observed for the IGF axis. While expression of Igf1 was downregulated, that of Igfbp1 was upregulated in the livers of both genders. Corresponding changes in the serum levels of both proteins could be detected by ELISA. By activating and inhibiting the transcriptional output of Hedgehog signaling in cultured hepatocytes through siRNAs against Ptch1 and Gli3, respectively, in combination with a ChIP assay evidence was collected indicating that Igf1 expression is directly dependent on the activator function of Gli3. In contrast, the mRNA level of Igfbp1 appears to be controlled through the repressor function of Gli3, while that of Igfbp2 and Igfbp3 did not change. Interestingly, body weight of the transgenic mice correlated well with IGF-I levels in both genders and also with IGFBP-1 levels in females, whereas it did not correlate with serum growth hormone levels.
Conclusions: Our results demonstrate for the first time that Hedgehog signaling is active in healthy mature mouse hepatocytes and that it has considerable importance for IGF-I homeostasis in the circulation. These findings may have various implications for mouse physiology including the regulation of body weight and size, glucose homeostasis and reproductive capacity.
Figures
References
-
- Ruiz I, Altaba A. Gli proteins encode context-dependent positive and negative functions: implications for development and disease. Development. 1999;126:3205–3216. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

