Cuckoo search epistasis: a new method for exploring significant genetic interactions
- PMID: 24549111
- PMCID: PMC4023449
- DOI: 10.1038/hdy.2014.4
Cuckoo search epistasis: a new method for exploring significant genetic interactions
Abstract
The advent of high-throughput sequencing technology has resulted in the ability to measure millions of single-nucleotide polymorphisms (SNPs) from thousands of individuals. Although these high-dimensional data have paved the way for better understanding of the genetic architecture of common diseases, they have also given rise to challenges in developing computational methods for learning epistatic relationships among genetic markers. We propose a new method, named cuckoo search epistasis (CSE) for identifying significant epistatic interactions in population-based association studies with a case-control design. This method combines a computationally efficient Bayesian scoring function with an evolutionary-based heuristic search algorithm, and can be efficiently applied to high-dimensional genome-wide SNP data. The experimental results from synthetic data sets show that CSE outperforms existing methods including multifactorial dimensionality reduction and Bayesian epistasis association mapping. In addition, on a real genome-wide data set related to Alzheimer's disease, CSE identified SNPs that are consistent with previously reported results, and show the utility of CSE for application to genome-wide data.
Figures



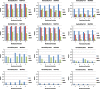
References
-
- Chung Y, Lee SY, Elston RC, Park T. Odds ratio based multifactor-dimensionality reduction method for detecting gene-gene interactions. Bioinformatics. 2007;23:71–76. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

