A Kinetic Model Explains Why Shorter and Less Affine Enzyme-recruiting Oligonucleotides Can Be More Potent
- PMID: 24549300
- PMCID: PMC3951909
- DOI: 10.1038/mtna.2013.72
A Kinetic Model Explains Why Shorter and Less Affine Enzyme-recruiting Oligonucleotides Can Be More Potent
Abstract
Antisense oligonucleotides complementary to RNA targets promise generality and ease of drug design. The first systemically administered antisense drug was recently approved for treatment and others are in clinical development. Chemical modifications that increase the hybridization affinity of oligonucleotides are reasoned to confer higher potency, i.e., modified oligonucleotides can be dosed at lower concentrations to achieve the same effect. Surprisingly, shorter and less affine oligonucleotides sometimes display increased potency. To explain this apparent contradiction, increased uptake or decreased propensity to form structures have been suggested as possible mechanisms. Here, we provide an alternative explanation that invokes only the kinetics behind oligonucleotide-mediated cleavage of RNA targets. A model based on the law of mass action predicts, and experiments support, the existence of an optimal binding affinity. Exaggerated affinity, and not length per se, is detrimental to potency. This finding clarifies how to optimally apply high-affinity modifications in the discovery of potent antisense oligonucleotide drugs.
Figures
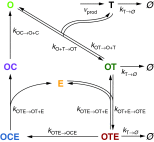
 . Here,
. Here,  denotes completely degraded target. The dissociation rate of enzyme from OT and OC is assumed to be the same.
denotes completely degraded target. The dissociation rate of enzyme from OT and OC is assumed to be the same.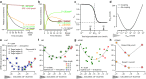
 and
and  . Solid line:
. Solid line:  . (e–g) Experimental knockdown as a function of calculated ΔG°: (e) for 21 oligonucleotides at 2 nmol/l targeted against the luciferase firefly gene, (f) for 14 oligonucleotides at 3 nmol/l targeted against the glucocorticoid receptor, and (g) for 23 oligonucleotides at 1 nmol/l targeted against apolipoprotein B (APOB), (h) 4 oligonucleotides at 0.06 μmol/l and 1.5 μmol/l against PCSK9. Legend indicates oligonucleotide lengths. Target messenger RNA concentrations are measured by (e) luciferase assay and (f,g,h) quantitative reverse transcriptase–polymerase chain reaction. Dots are experimental data and gray lines are a least squares fit to a second-order polynomial with P values and vertex for the fit in each of the panels e–h.
. (e–g) Experimental knockdown as a function of calculated ΔG°: (e) for 21 oligonucleotides at 2 nmol/l targeted against the luciferase firefly gene, (f) for 14 oligonucleotides at 3 nmol/l targeted against the glucocorticoid receptor, and (g) for 23 oligonucleotides at 1 nmol/l targeted against apolipoprotein B (APOB), (h) 4 oligonucleotides at 0.06 μmol/l and 1.5 μmol/l against PCSK9. Legend indicates oligonucleotide lengths. Target messenger RNA concentrations are measured by (e) luciferase assay and (f,g,h) quantitative reverse transcriptase–polymerase chain reaction. Dots are experimental data and gray lines are a least squares fit to a second-order polynomial with P values and vertex for the fit in each of the panels e–h.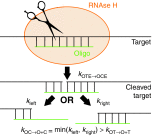
 . Upon enzyme (E, RNase H) binding to the OT complex the target is cleaved at a rate
. Upon enzyme (E, RNase H) binding to the OT complex the target is cleaved at a rate  . After cleavage, the right and left parts of the cleaved target will dissociate from the oligonucleotide at (faster) rates kright and kleft, respectively.
. After cleavage, the right and left parts of the cleaved target will dissociate from the oligonucleotide at (faster) rates kright and kleft, respectively.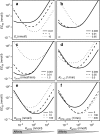
 , (e) the dissociation constant for the OTE complex, KdOTE, and (f) the rate of target cleavage,
, (e) the dissociation constant for the OTE complex, KdOTE, and (f) the rate of target cleavage,  .
.References
-
- Stein H, Hausen P. Enzyme from calf thymus degrading the RNA moiety of DNA-RNA Hybrids: effect on DNA-dependent RNA polymerase. Science. 1969;166:393–395. - PubMed
-
- Bennett CF, Swayze EE. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu Rev Pharmacol Toxicol. 2010;50:259–293. - PubMed
-
- Obika S, Nanbu D, Hari Y, Morio K, In Y, Ishida T, et al. Synthesis of 2-O,4-C-methyleneuridine and -cytidine. Novel bicyclic nucleosides having a fixed C3, -endo sugar puckering. Tet Lett. 1997;38:8735–8738.
LinkOut - more resources
Full Text Sources
Other Literature Sources

