Reovirus forms neo-organelles for progeny particle assembly within reorganized cell membranes
- PMID: 24549844
- PMCID: PMC3944815
- DOI: 10.1128/mBio.00931-13
Reovirus forms neo-organelles for progeny particle assembly within reorganized cell membranes
Erratum in
-
Correction for Fernández de Castro et al., Reovirus Forms Neo-Organelles for Progeny Particle Assembly within Reorganized Cell Membranes.mBio. 2015 Jan 27;6(1):e02529-14. doi: 10.1128/mBio.02529-14. mBio. 2015. PMID: 25626908 Free PMC article. No abstract available.
Abstract
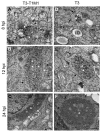
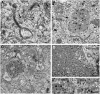
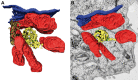
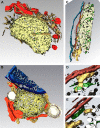
Most viruses that replicate in the cytoplasm of host cells form neo-organelles that serve as sites of viral genome replication and particle assembly. These highly specialized structures concentrate viral replication proteins and nucleic acids, prevent the activation of cell-intrinsic defenses, and coordinate the release of progeny particles. Despite the importance of inclusion complexes in viral replication, there are key gaps in the knowledge of how these organelles form and mediate their functions. Reoviruses are nonenveloped, double-stranded RNA (dsRNA) viruses that serve as tractable experimental models for studies of dsRNA virus replication and pathogenesis. Following reovirus entry into cells, replication occurs in large cytoplasmic structures termed inclusions that fill with progeny virions. Reovirus inclusions are nucleated by viral nonstructural proteins, which in turn recruit viral structural proteins for genome replication and particle assembly. Components of reovirus inclusions are poorly understood, but these structures are generally thought to be devoid of membranes. We used transmission electron microscopy and three-dimensional image reconstructions to visualize reovirus inclusions in infected cells. These studies revealed that reovirus inclusions form within a membranous network. Viral inclusions contain filled and empty viral particles and microtubules and appose mitochondria and rough endoplasmic reticulum (RER). Immunofluorescence confocal microscopy analysis demonstrated that markers of the ER and ER-Golgi intermediate compartment (ERGIC) codistribute with inclusions during infection, as does dsRNA. dsRNA colocalizes with the viral protein σNS and an ERGIC marker inside inclusions. These findings suggest that cell membranes within reovirus inclusions form a scaffold to coordinate viral replication and assembly.
Importance: Viruses alter the architecture of host cells to form an intracellular environment conducive to viral replication. This step in viral infection requires the concerted action of viral and host components and is potentially vulnerable to pharmacological intervention. Reoviruses form large cytoplasmic replication sites called inclusions, which have been described as membrane-free structures. Despite the importance of inclusions in the reovirus replication cycle, little is known about their formation and composition. We used light and electron microscopy to demonstrate that reovirus inclusions are membrane-containing structures and that the endoplasmic reticulum (ER) and the ER-Golgi intermediate compartment interact closely with these viral organelles. These findings enhance our understanding of the cellular machinery usurped by viruses to form inclusion organelles and complete an infectious cycle. This information, in turn, may foster the development of antiviral drugs that impede this essential viral replication step.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
