Systems biology-based identification of Mycobacterium tuberculosis persistence genes in mouse lungs
- PMID: 24549847
- PMCID: PMC3944818
- DOI: 10.1128/mBio.01066-13
Systems biology-based identification of Mycobacterium tuberculosis persistence genes in mouse lungs
Abstract
Identifying Mycobacterium tuberculosis persistence genes is important for developing novel drugs to shorten the duration of tuberculosis (TB) treatment. We developed computational algorithms that predict M. tuberculosis genes required for long-term survival in mouse lungs. As the input, we used high-throughput M. tuberculosis mutant library screen data, mycobacterial global transcriptional profiles in mice and macrophages, and functional interaction networks. We selected 57 unique, genetically defined mutants (18 previously tested and 39 untested) to assess the predictive power of this approach in the murine model of TB infection. We observed a 6-fold enrichment in the predicted set of M. tuberculosis genes required for persistence in mouse lungs relative to randomly selected mutant pools. Our results also allowed us to reclassify several genes as required for M. tuberculosis persistence in vivo. Finally, the new results implicated additional high-priority candidate genes for testing. Experimental validation of computational predictions demonstrates the power of this systems biology approach for elucidating M. tuberculosis persistence genes.
Importance: Mycobacterium tuberculosis, the causative agent of tuberculosis (TB), has a genetic repertoire that permits it to persist in the face of host immune responses. Identification of such persistence genes could reveal novel drug targets and elucidate mechanisms by which the organism eludes the immune system and resists drugs. Genetic screens have identified a total of 31 persistence genes, but to date only 15% of the ~4,000 M. tuberculosis genes have been tested experimentally. In this paper, as an alternative to brute force experimental screens, we describe computational methods that predict new persistence genes by combining known examples with growing databases of biological networks. Experimental testing demonstrated that these predictions are highly accurate, validating the computational approach and providing new information about M. tuberculosis persistence in host tissues. Using the new experimental results as additional input highlights additional genes for testing. Our approach can be extended to other data types and target organisms to characterize host-pathogen interactions relevant to this and other infectious diseases.
Figures


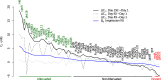
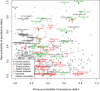
References
-
- Lamichhane G, Zignol M, Blades NJ, Geiman DE, Dougherty A, Grosset J, Broman KW, Bishai WR. 2003. A postgenomic method for predicting essential genes at subsaturation levels of mutagenesis: application to Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 100:7213–7218. 10.1073/pnas.1231432100 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
