The bHLH transcription factor HBI1 mediates the trade-off between growth and pathogen-associated molecular pattern-triggered immunity in Arabidopsis
- PMID: 24550223
- PMCID: PMC3967043
- DOI: 10.1105/tpc.113.121111
The bHLH transcription factor HBI1 mediates the trade-off between growth and pathogen-associated molecular pattern-triggered immunity in Arabidopsis
Abstract
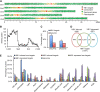
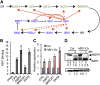
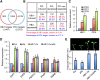
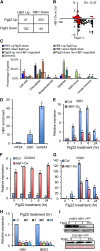
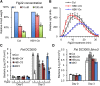
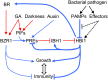
The trade-off between growth and immunity is crucial for survival in plants. However, the mechanism underlying growth-immunity balance has remained elusive. The PRE-IBH1-HBI1 tripartite helix-loop-helix/basic helix-loop-helix module is part of a central transcription network that mediates growth regulation by several hormonal and environmental signals. Here, genome-wide analyses of HBI1 target genes show that HBI1 regulates both overlapping and unique targets compared with other DNA binding components of the network in Arabidopsis thaliana, supporting a role in specifying network outputs and fine-tuning feedback regulation. Furthermore, HBI1 negatively regulates a subset of genes involved in immunity, and pathogen-associated molecular pattern (PAMP) signals repress HBI1 transcription. Constitutive overexpression and loss-of-function experiments show that HBI1 inhibits PAMP-induced growth arrest, defense gene expression, reactive oxygen species production, and resistance to pathogen. These results show that HBI1, as a component of the central growth regulation circuit, functions as a major node of crosstalk that mediates a trade-off between growth and immunity in plants.
Figures
References
-
- Albrecht C., Boutrot F., Segonzac C., Schwessinger B., Gimenez-Ibanez S., Chinchilla D., Rathjen J.P., de Vries S.C., Zipfel C. (2012). Brassinosteroids inhibit pathogen-associated molecular pattern-triggered immune signaling independent of the receptor kinase BAK1. Proc. Natl. Acad. Sci. USA 109: 303–308. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

