Apyrase suppression raises extracellular ATP levels and induces gene expression and cell wall changes characteristic of stress responses
- PMID: 24550243
- PMCID: PMC3982762
- DOI: 10.1104/pp.113.233429
Apyrase suppression raises extracellular ATP levels and induces gene expression and cell wall changes characteristic of stress responses
Abstract
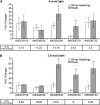
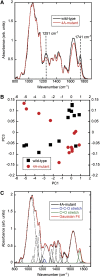
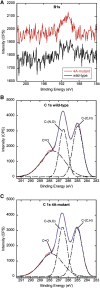
Plant cells release ATP into their extracellular matrix as they grow, and extracellular ATP (eATP) can modulate the rate of cell growth in diverse tissues. Two closely related apyrases (APYs) in Arabidopsis (Arabidopsis thaliana), APY1 and APY2, function, in part, to control the concentration of eATP. The expression of APY1/APY2 can be inhibited by RNA interference, and this suppression leads to an increase in the concentration of eATP in the extracellular medium and severely reduces growth. To clarify how the suppression of APY1 and APY2 is linked to growth inhibition, the gene expression changes that occur in seedlings when apyrase expression is suppressed were assayed by microarray and quantitative real-time-PCR analyses. The most significant gene expression changes induced by APY suppression were in genes involved in biotic stress responses, which include those genes regulating wall composition and extensibility. These expression changes predicted specific chemical changes in the walls of mutant seedlings, and two of these changes, wall lignification and decreased methyl ester bonds, were verified by direct analyses. Taken together, the results are consistent with the hypothesis that APY1, APY2, and eATP play important roles in the signaling steps that link biotic stresses to plant defense responses and growth changes.
Figures




References
-
- Almagro L, Gómez Ros LV, Belchi-Navarro S, Bru R, Ros Barceló A, Pedreño MA. (2009) Class III peroxidases in plant defence reactions. J Exp Bot 60: 377–390 - PubMed
-
- Aquea F, Federici F, Moscoso C, Vega A, Jullian P, Haseloff J, Arce-Johnson P. (2012) A molecular framework for the inhibition of Arabidopsis root growth in response to boron toxicity. Plant Cell Environ 35: 719–734 - PubMed
-
- Bouwmeester K, Han M, Blanco-Portales R, Song W, Weide R, Guo LY, van der Vossen EAG, Govers F. (2014) The Arabidopsis lectin receptor kinase LecRK-I.9 enhances resistance to Phytophthora infestans in Solanaceous plants. Plant Biotechnol J 12: 10–16 - PubMed
-
- Chen YF, Shakeel SN, Bowers J, Zhao XC, Etheridge N, Schaller GE. (2007) Ligand-induced degradation of the ethylene receptor ETR2 through a proteasome-dependent pathway in Arabidopsis. J Biol Chem 282: 24752–24758 - PubMed
-
- Chiu TY, Christiansen K, Moreno I, Lao JM, Loqué D, Orellana A, Heazlewood JL, Clark G, Roux SJ. (2012) AtAPY1 and AtAPY2 function as Golgi-localized nucleoside diphosphatases in Arabidopsis thaliana. Plant Cell Physiol 53: 1913–1925 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

