miR-146a promotes the initiation and progression of melanoma by activating Notch signaling
- PMID: 24550252
- PMCID: PMC3927633
- DOI: 10.7554/eLife.01460
miR-146a promotes the initiation and progression of melanoma by activating Notch signaling
Abstract
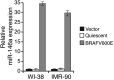
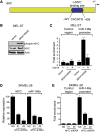
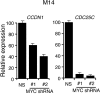
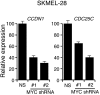
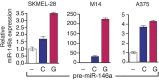
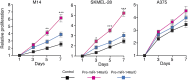
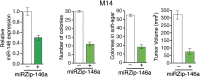
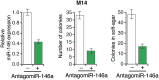
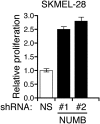
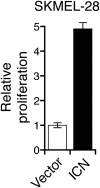
Oncogenic mutations in BRAF and NRAS occur in 70% of melanomas. In this study, we identify a microRNA, miR-146a, that is highly upregulated by oncogenic BRAF and NRAS. Expression of miR-146a increases the ability of human melanoma cells to proliferate in culture and form tumors in mice, whereas knockdown of miR-146a has the opposite effects. We show these oncogenic activities are due to miR-146a targeting the NUMB mRNA, a repressor of Notch signaling. Previous studies have shown that pre-miR-146a contains a single nucleotide polymorphism (C>G rs2910164). We find that the ability of pre-miR-146a/G to activate Notch signaling and promote oncogenesis is substantially higher than that of pre-miR-146a/C. Analysis of melanoma cell lines and matched patient samples indicates that during melanoma progression pre-miR-146a/G is enriched relative to pre-miR-146a/C, resulting from a C-to-G somatic mutation in pre-miR-146a/C. Collectively, our results reveal a central role for miR-146a in the initiation and progression of melanoma. DOI: http://dx.doi.org/10.7554/eLife.01460.001.
Keywords: BRAF; NRAS; melanoma; miRNA.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures
















References
-
- Aubin-Houzelstein G, Djian-Zaouche J, Bernex F, Gadin S, Delmas V, Larue L, Panthier JJ. 2008. Melanoblasts’ proper location and timed differentiation depend on Notch/RBP-J signaling in postnatal hair follicles. The Journal of Investigative Dermatology 128:2686–2695. 10.1038/jid.2008.120 - DOI - PubMed
-
- Balint K, Xiao M, Pinnix CC, Soma A, Veres I, Juhasz I, Brown EJ, Capobianco AJ, Herlyn M, Liu ZJ. 2005. Activation of Notch1 signaling is required for beta-catenin-mediated human primary melanoma progression. The Journal of Clinical Investigation 115:3166–3176. 10.1172/JCI25001 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

