Reciprocal interactions of human C10orf12 and C17orf96 with PRC2 revealed by BioTAP-XL cross-linking and affinity purification
- PMID: 24550272
- PMCID: PMC3932854
- DOI: 10.1073/pnas.1400648111
Reciprocal interactions of human C10orf12 and C17orf96 with PRC2 revealed by BioTAP-XL cross-linking and affinity purification
Abstract
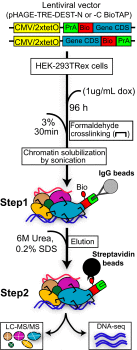
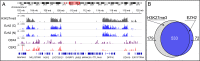
Understanding the composition of epigenetic regulators remains an important challenge in chromatin biology. Traditional biochemical analysis of chromatin-associated complexes requires their release from DNA under conditions that can also disrupt key interactions. Here we develop a complementary approach (BioTAP-XL), in which cross-linking (XL) enhances the preservation of protein interactions and also allows the analysis of DNA targets under the same tandem affinity purification (BioTAP) regimen. We demonstrate the power of BioTAP-XL through analysis of human EZH2, a core subunit of polycomb repressive complex 2 (PRC2). We identify and validate two strong interactors, C10orf12 and C17orf96, which display enrichment with EZH2-BioTAP at levels similar to canonical PRC2 components (SUZ12, EED, MTF2, JARID2, PHF1, and AEBP2). ChIP-seq analysis of BioTAP-tagged C10orf12 or C17orf96 revealed the similarity of each binding pattern with the location of EZH2 and the H3K27me3-silencing mark, validating their physical interaction with PRC2 components. Interestingly, analysis by mass spectrometry of C10orf12 and C17orf96 interactions revealed that these proteins may be mutually exclusive PRC2 subunits that fail to interact with each other or with JARID2 and AEBP2. C10orf12, in addition, shows a strong and unexpected association with components of the EHMT1/2 complex, thus potentially connecting PRC2 to another histone methyltransferase. Similarly, results from CBX4-BioTAP protein pulldowns are consistent with reports of a diversity of PRC1 complexes. Our results highlight the importance of reciprocal analyses of multiple subunits and suggest that iterative use of BioTAP-XL has strong potential to reveal networks of chromatin-based interactions in higher organisms.
Keywords: LC-MS/MS; chromatin IP; formaldehyde cross-linking; protein–protein interactions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

