The tripartite motif coiled-coil is an elongated antiparallel hairpin dimer
- PMID: 24550273
- PMCID: PMC3932864
- DOI: 10.1073/pnas.1318962111
The tripartite motif coiled-coil is an elongated antiparallel hairpin dimer
Abstract
Tripartite motif (TRIM) proteins make up a large family of coiled-coil-containing RING E3 ligases that function in many cellular processes, particularly innate antiviral response pathways. Both dimerization and higher-order assembly are important elements of TRIM protein function, but the atomic details of TRIM tertiary and quaternary structure have not been fully understood. Here, we present crystallographic and biochemical analyses of the TRIM coiled-coil and show that TRIM proteins dimerize by forming interdigitating antiparallel helical hairpins that position the N-terminal catalytic RING domains at opposite ends of the dimer and the C-terminal substrate-binding domains at the center. The dimer core comprises an antiparallel coiled-coil with a distinctive, symmetric pattern of flanking heptad and central hendecad repeats that appear to be conserved across the entire TRIM family. Our studies reveal how the coiled-coil organizes TRIM25 to polyubiquitylate the RIG-I/viral RNA recognition complex and how dimers of the TRIM5α protein are arranged within hexagonal arrays that recognize the HIV-1 capsid lattice and restrict retroviral replication.
Keywords: X-ray crystallography; antiparallel dimer; disulfide crosslinking.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
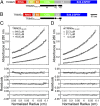
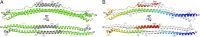

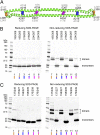
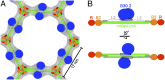
References
-
- McNab FW, Rajsbaum R, Stoye JP, O’Garra A. Tripartite-motif proteins and innate immune regulation. Curr Opin Immunol. 2011;23(1):46–56. - PubMed
-
- Gack MU, et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446(7138):916–920. - PubMed
-
- Oshiumi H, Matsumoto M, Seya T. Ubiquitin-mediated modulation of the cytoplasmic viral RNA sensor RIG-I. J Biochem. 2012;151(1):5–11. - PubMed
-
- Stremlau M, et al. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature. 2004;427(6977):848–853. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

