Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission
- PMID: 24550281
- PMCID: PMC3932921
- DOI: 10.1073/pnas.1324297111
Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission
Abstract
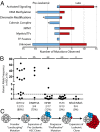
Cancer is widely characterized by the sequential acquisition of genetic lesions in a single lineage of cells. Our previous studies have shown that, in acute myeloid leukemia (AML), mutation acquisition occurs in functionally normal hematopoietic stem cells (HSCs). These preleukemic HSCs harbor some, but not all, of the mutations found in the leukemic cells. We report here the identification of patterns of mutation acquisition in human AML. Our findings support a model in which mutations in "landscaping" genes, involved in global chromatin changes such as DNA methylation, histone modification, and chromatin looping, occur early in the evolution of AML, whereas mutations in "proliferative" genes occur late. Additionally, we analyze the persistence of preleukemic mutations in patients in remission and find CD34+ progenitor cells and various mature cells that harbor preleukemic mutations. These findings indicate that preleukemic HSCs can survive induction chemotherapy, identifying these cells as a reservoir for the reevolution of relapsed disease. Finally, through the study of several cases of relapsed AML, we demonstrate various evolutionary patterns for the generation of relapsed disease and show that some of these patterns are consistent with involvement of preleukemic HSCs. These findings provide key insights into the monitoring of minimal residual disease and the identification of therapeutic targets in human AML.
Keywords: clonal evolution; preleukemia.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Comment in
-
On the origin of leukemic species.Cell Stem Cell. 2014 Apr 3;14(4):421-2. doi: 10.1016/j.stem.2014.03.008. Cell Stem Cell. 2014. PMID: 24702991
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

