Loss of the tumor suppressor LKB1 promotes metabolic reprogramming of cancer cells via HIF-1α
- PMID: 24550282
- PMCID: PMC3932920
- DOI: 10.1073/pnas.1312570111
Loss of the tumor suppressor LKB1 promotes metabolic reprogramming of cancer cells via HIF-1α
Abstract
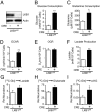
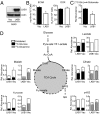
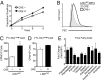
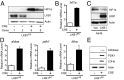
One of the major metabolic changes associated with cellular transformation is enhanced nutrient utilization, which supports tumor progression by fueling both energy production and providing biosynthetic intermediates for growth. The liver kinase B1 (LKB1) is a serine/threonine kinase and tumor suppressor that couples bioenergetics to cell-growth control through regulation of mammalian target of rapamycin (mTOR) activity; however, the influence of LKB1 on tumor metabolism is not well defined. Here, we show that loss of LKB1 induces a progrowth metabolic program in proliferating cells. Cells lacking LKB1 display increased glucose and glutamine uptake and utilization, which support both cellular ATP levels and increased macromolecular biosynthesis. This LKB1-dependent reprogramming of cell metabolism is dependent on the hypoxia-inducible factor-1α (HIF-1α), which accumulates under normoxia in LKB1-deficient cells and is antagonized by inhibition of mTOR complex I signaling. Silencing HIF-1α reverses the metabolic advantages conferred by reduced LKB1 signaling and impairs the growth and survival of LKB1-deficient tumor cells under low-nutrient conditions. Together, our data implicate the tumor suppressor LKB1 as a central regulator of tumor metabolism and growth control through the regulation of HIF-1α-dependent metabolic reprogramming.
Keywords: HIF-1alpha; PJS; Peutz-Jeghers Syndrome; Warburg effect; cancer metabolism; glutamine metabolism.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
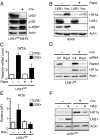

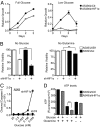
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

