Trimethylamine N-oxide metabolism by abundant marine heterotrophic bacteria
- PMID: 24550299
- PMCID: PMC3932874
- DOI: 10.1073/pnas.1317834111
Trimethylamine N-oxide metabolism by abundant marine heterotrophic bacteria
Abstract
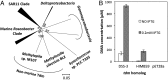
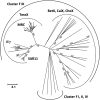
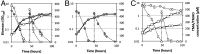
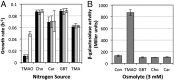
Trimethylamine N-oxide (TMAO) is a common osmolyte found in a variety of marine biota and has been detected at nanomolar concentrations in oceanic surface waters. TMAO can serve as an important nutrient for ecologically important marine heterotrophic bacteria, particularly the SAR11 clade and marine Roseobacter clade (MRC). However, the enzymes responsible for TMAO catabolism and the membrane transporter required for TMAO uptake into microbial cells have yet to be identified. We show here that the enzyme TMAO demethylase (Tdm) catalyzes the first step in TMAO degradation. This enzyme represents a large group of proteins with an uncharacterized domain (DUF1989). The function of TMAO demethylase in a representative from the SAR11 clade (strain HIMB59) and in a representative of the MRC (Ruegeria pomeroyi DSS-3) was confirmed by heterologous expression of tdm (the gene encoding Tdm) in Escherichia coli. In R. pomeroyi, mutagenesis experiments confirmed that tdm is essential for growth on TMAO. We also identified a unique ATP-binding cassette transporter (TmoXWV) found in a variety of marine bacteria and experimentally confirmed its specificity for TMAO through marker exchange mutagenesis and lacZ reporter assays of the promoter for genes encoding this transporter. Both Tdm and TmoXWV are particularly abundant in natural seawater assemblages and actively expressed, as indicated by a number of recent metatranscriptomic and metaproteomic studies. These data suggest that TMAO represents a significant, yet overlooked, nutrient for marine bacteria.
Keywords: TMAO transporter; marine nitrogen cycle; methylated amine metabolism; nitrogenous osmolyte.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Yancey PH, Clark ME, Hand SC, Bowlus RD, Somero GN. Living with water stress: Evolution of osmolyte systems. Science. 1982;217(4566):1214–1222. - PubMed
-
- Seibel BA, Walsh PJ. Trimethylamine oxide accumulation in marine animals: Relationship to acylglycerol storage. J Exp Biol. 2002;205(Pt 3):297–306. - PubMed
-
- Ballantyne JS. Jaws: The inside story. the metabolism of elasmobranch fishes. Comp Biochem Physiol B Biochem Mol Biol. 1997;118(4):703–742.
-
- Treberg JR, et al. The accumulation of methylamine counteracting solutes in elasmobranchs with differing levels of urea: A comparison of marine and freshwater species. J Exp Biol. 2006;209(Pt 5):860–870. - PubMed
-
- Quinn PK, Charlson RJ, Bates TS. Simultaneous observations of ammonia in the atmosphere and ocean. Nature. 1988;335(6188):336–338.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

