Identification of a conserved branched RNA structure that functions as a factor-independent terminator
- PMID: 24550474
- PMCID: PMC3948284
- DOI: 10.1073/pnas.1315374111
Identification of a conserved branched RNA structure that functions as a factor-independent terminator
Abstract
Anti-Q is a small RNA encoded on pCF10, an antibiotic resistance plasmid of Enterococcus faecalis, which negatively regulates conjugation of the plasmid. In this study we sought to understand how Anti-Q is generated relative to larger transcripts of the same operon. We found that Anti-Q folds into a branched structure that functions as a factor-independent terminator. In vitro and in vivo, termination is dependent on the integrity of this structure as well as the presence of a 3' polyuridine tract, but is not dependent on other downstream sequences. In vitro, terminated transcripts are released from RNA polymerase after synthesis. In vivo, a mutant with reduced termination efficiency demonstrated loss of tight control of conjugation function. A search of bacterial genomes revealed the presence of sequences that encode Anti-Q-like RNA structures. In vitro and in vivo experiments demonstrated that one of these functions as a terminator. This work reveals a previously unappreciated flexibility in the structure of factor-independent terminators and identifies a mechanism for generation of functional small RNAs; it should also inform annotation of bacterial sequence features, such as terminators, functional sRNAs, and operons.
Keywords: antisense; attenuation; cell–cell signaling; pheromone; transcription.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


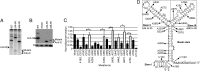

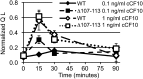
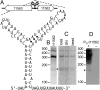
References
-
- Nudler E, Gottesman ME. Transcription termination and anti-termination in E. coli. Genes Cells. 2002;7(8):755–768. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

