Gene expression differences underlying genotype-by-genotype specificity in a host-parasite system
- PMID: 24550506
- PMCID: PMC3948297
- DOI: 10.1073/pnas.1318628111
Gene expression differences underlying genotype-by-genotype specificity in a host-parasite system
Abstract
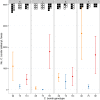
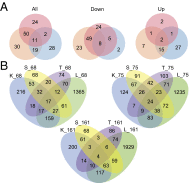
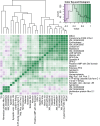
In many systems, host-parasite evolutionary dynamics have led to the emergence and maintenance of diverse parasite and host genotypes within the same population. Genotypes vary in key attributes: Parasite genotypes vary in ability to infect, host genotypes vary in susceptibility, and infection outcome is frequently the result of both parties' genotypic identities. These host-parasite genotype-by-genotype (GH × GP) interactions influence evolutionary and ecological dynamics in important ways. Interactions can be produced through genetic variation; however, here, we assess the role of variable gene expression as an additional source of GH × GP interactions. The bumblebee Bombus terrestris and its trypanosome gut parasite Crithidia bombi are a model system for host-parasite matching. Full-transcriptome sequencing of the bumblebee host revealed that different parasite genotypes indeed induce fundamentally different host expression responses and host genotypes vary in their responses to the infecting parasite genotype. It appears that broadly and successfully infecting parasite genotypes lead to reduced host immune gene expression relative to unexposed bees but induce the expression of genes responsible for controlling gene expression. Contrastingly, a poorly infecting parasite genotype induced the expression of immunologically important genes, including antimicrobial peptides. A targeted expression assay confirmed the transcriptome results and also revealed strong host genotype effects. In all, the expression of a number of genes depends on the host genotype and the parasite genotype and the interaction between both host and parasite genotypes. These results suggest that alongside sequence variation in coding immunological genes, variation that controls immune gene expression can also produce patterns of host-parasite specificity.
Keywords: Red Queen; coevolution; manipulation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Carius HJ, Little TJ, Ebert D. Genetic variation in a host-parasite association: Potential for coevolution and frequency-dependent selection. Evolution. 2001;55(6):1136–1145. - PubMed
-
- Schmid-Hempel P. On the evolutionary ecology of host-parasite interactions: Addressing the question with regard to bumblebees and their parasites. Naturwissenschaften. 2001;88(4):147–158. - PubMed
-
- Mallon EB, Loosli R, Schmid-Hempel P. Specific versus nonspecific immune defense in the bumblebee, Bombus terrestris L. Evolution. 2003;57(6):1444–1447. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

