Significant role of PB1 and UBA domains in multimerization of Joka2, a selective autophagy cargo receptor from tobacco
- PMID: 24550923
- PMCID: PMC3907767
- DOI: 10.3389/fpls.2014.00013
Significant role of PB1 and UBA domains in multimerization of Joka2, a selective autophagy cargo receptor from tobacco
Abstract
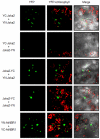
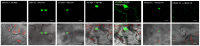
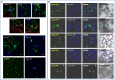
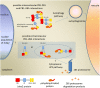
Tobacco Joka2 protein is a hybrid homolog of two mammalian selective autophagy cargo receptors, p62 and NBR1. These proteins can directly interact with the members of ATG8 family and the polyubiquitinated cargoes designed for degradation. Function of the selective autophagy cargo receptors relies on their ability to form protein aggregates. It has been shown that the N-terminal PB1 domain of p62 is involved in formation of aggregates, while the UBA domains of p62 and NBR1 have been associated mainly with cargo binding. Here we focus on roles of PB1 and UBA domains in localization and aggregation of Joka2 in plant cells. We show that Joka2 can homodimerize not only through its N-terminal PB1-PB1 interactions but also via interaction between N-terminal PB1 and C-terminal UBA domains. We also demonstrate that Joka2 co-localizes with recombinant ubiquitin and sequestrates it into aggregates and that C-terminal part (containing UBA domains) is sufficient for this effect. Our results indicate that Joka2 accumulates in cytoplasmic aggregates and suggest that in addition to these multimeric forms it also exists in the nucleus and cytoplasm in a monomeric form.
Keywords: Joka2; NBR1; PB1; UBA; autophagy; proteasome; selective autophagy cargo receptor; ubiquitin.
Figures





References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

