Effect of Target Length on Specificity and Sensitivity of Oligonucleotide Microarrays: A Comparison between Dendrimer and Modified PCR based Labelling Methods
- PMID: 24551024
- PMCID: PMC3927376
- DOI: 10.2174/1874091X01408010011
Effect of Target Length on Specificity and Sensitivity of Oligonucleotide Microarrays: A Comparison between Dendrimer and Modified PCR based Labelling Methods
Abstract
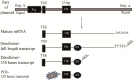
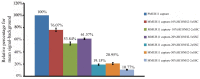
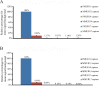
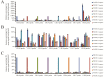
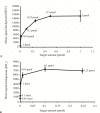
DNA microarrays are widely used as end point detectors for gene expression analysis. Several methods have been developed for target labelling to enable quantification but without taking target length into consideration. Here we highlight the importance of choosing the optimum target length that would ensure specificity without compromising sensitivity of the assay. For this, eight plasmids that are identical to each other except for a closely related 23 bp unique reporter (UR) sequence were used to examine the hybridization efficiency for these URs. Targets of various lengths were generated and labelled as follows: full length and 330 bases transcripts using a dendrimer labelling method, 120 bp amplicons by the modified PCR end labelling method and synthetic labelled targets of 33 bases. This report also shows the advantages of using the modified PCR method over other labelling methods in generating labelled amplicons of the desired lengths to maximize hybridization efficiency.
Keywords: Dendrimer.; Hybridization specificity; Oligonucleotide microarrays; PCR; Secondary structure; Sensitivity.
Figures
References
-
- Pollack JR, Perou CM, Alizadeh AA, Eisen MB, Pergamenschikov A, Williams CF, Jeffrey SS, Botstein D, Brown PO. Genomewide analysis of DNA copy-number changes using cDNA microarrays. Nat. Genet. 1999;23:41–46. - PubMed
-
- DeRisi JL, Iyer VR, Brown BO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. - PubMed
-
- Drmanac S, Kita D, Labat I, Hauser B, Schmidt C, Burczak JD, Drmanac R. Accurate sequencing by hybridization for DNA diagnostics and individual genomics. Nat. Biotechnol. 1998;16(1):54–58. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
