Variation, evolution, and correlation analysis of C+G content and genome or chromosome size in different kingdoms and phyla
- PMID: 24551092
- PMCID: PMC3923770
- DOI: 10.1371/journal.pone.0088339
Variation, evolution, and correlation analysis of C+G content and genome or chromosome size in different kingdoms and phyla
Abstract
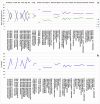
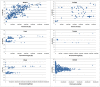
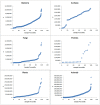
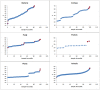
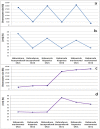
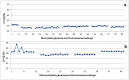
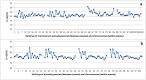
C+G content (GC content or G+C content) is known to be correlated with genome/chromosome size in bacteria but the relationship for other kingdoms remains unclear. This study analyzed genome size, chromosome size, and base composition in most of the available sequenced genomes in various kingdoms. Genome size tends to increase during evolution in plants and animals, and the same is likely true for bacteria. The genomic C+G contents were found to vary greatly in microorganisms but were quite similar within each animal or plant subkingdom. In animals and plants, the C+G contents are ranked as follows: monocot plants>mammals>non-mammalian animals>dicot plants. The variation in C+G content between chromosomes within species is greater in animals than in plants. The correlation between average chromosome C+G content and chromosome length was found to be positive in Proteobacteria, Actinobacteria (but not in other analyzed bacterial phyla), Ascomycota fungi, and likely also in some plants; negative in some animals, insignificant in two protist phyla, and likely very weak in Archaea. Clearly, correlations between C+G content and chromosome size can be positive, negative, or not significant depending on the kingdoms/groups or species. Different phyla or species exhibit different patterns of correlation between chromosome-size and C+G content. Most chromosomes within a species have a similar pattern of variation in C+G content but outliers are common. The data presented in this study suggest that the C+G content is under genetic control by both trans- and cis- factors and that the correlation between C+G content and chromosome length can be positive, negative, or not significant in different phyla.
Conflict of interest statement
Figures


References
-
- Chargaff E, Lipshitz R, Green C (1952) Composition of the desoxypentose nucleic acids of four genera of sea-urchin. J Biol Chem 195: 155–160. - PubMed
-
- Watson JD, Crick FHC (1953) Molecular structure of nucleic acids: A structure for deoxyribose nucleic acid. Nature 171: 737–738. - PubMed
-
- Cionini PG, Bassi P, Cremonini R, Cavallini A (1985) Cytological localization of fast renaturing and satellite DNA sequences in Vicia faba . Protoplasma 124: 106–111.
-
- Appels R, Dennis ES, Smyth DR, Peacock WJ (1981) Two repeated DNA sequences from the heterochromatic regions of rye (Secale cereale) chromosomes. Chromosoma 84: 265–277.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

