Identification and functional characterization of two executioner caspases in Crassostrea gigas
- PMID: 24551213
- PMCID: PMC3923871
- DOI: 10.1371/journal.pone.0089040
Identification and functional characterization of two executioner caspases in Crassostrea gigas
Abstract
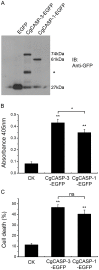
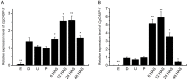
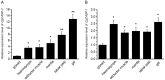
Caspase-3 and caspase-7 are two key effector caspases that play important roles in apoptotic pathways that maintain normal tissue and organ development and homeostasis. However, little is known about the sequence, structure, activity, and function of effector caspases upon apoptosis in mollusks, especially marine bivalves. In this study, we investigated the possible roles of two executioner caspases in the regulation of apoptosis in the Pacific oyster Crassostrea gigas. A full-length caspase-3-like gene named Cgcaspase-3 was cloned from C.gigas cDNA, encoding a predicted protein containing caspase family p20 and p10 domain profiles and a conserved caspase active site motif. Phylogenetic analysis demonstrated that both Cgcaspase-3 and Cgcaspase-1 may function as effector caspases clustered in the invertebrate branch. Although the sequence identities between the two caspases was low, both enzymes possessed executioner caspase activity and were capable of inducing cell death. These results suggested that Cgcaspase-3 and Cgcaspase-1 were two effector caspases in C. gigas. We also observed that nucleus-localized Cgcaspase-3, may function as a caspase-3-like protein and cytoplasm-localized Cgcaspase-1 may function as a caspase-7-like protein. Both Cgcaspase-3 and Cgcaspase-1 mRNA expression increased after larvae settled on the substratum, suggesting that both caspases acted in several tissues or organs that degenerated after oyster larvae settlement. The highest caspase expression levels were observed in the gills indicating that both effector caspases were likely involved in immune or metabolic processes in C. gigas.
Conflict of interest statement
Figures




Similar articles
-
The self-activation and LPS binding activity of executioner caspase-1 in oyster Crassostrea gigas.Dev Comp Immunol. 2017 Dec;77:330-339. doi: 10.1016/j.dci.2017.09.002. Epub 2017 Sep 6. Dev Comp Immunol. 2017. PMID: 28888538
-
Cloning and characterization of a novel caspase-8-like gene in Crassostrea gigas.Fish Shellfish Immunol. 2015 Oct;46(2):486-92. doi: 10.1016/j.fsi.2015.06.035. Epub 2015 Jul 2. Fish Shellfish Immunol. 2015. PMID: 26143079
-
Characterization and expression of a novel caspase gene: Evidence of the expansion of caspases in Crassostrea gigas.Comp Biochem Physiol B Biochem Mol Biol. 2016 Nov;201:37-45. doi: 10.1016/j.cbpb.2016.07.001. Epub 2016 Jul 6. Comp Biochem Physiol B Biochem Mol Biol. 2016. PMID: 27393814
-
Bacterial secreted effectors and caspase-3 interactions.Cell Microbiol. 2014 Dec;16(12):1746-56. doi: 10.1111/cmi.12368. Epub 2014 Oct 30. Cell Microbiol. 2014. PMID: 25262664 Free PMC article. Review.
-
Expansion and diversity of caspases in Mytilus coruscus contribute to larval metamorphosis and environmental adaptation.BMC Genomics. 2024 Mar 27;25(1):314. doi: 10.1186/s12864-024-10238-w. BMC Genomics. 2024. PMID: 38532358 Free PMC article. Review.
Cited by
-
Molecular cloning of two molluscan caspases and gene functional analysis during Crassostrea angulata (Fujian oyster) larval metamorphosis.Mol Biol Rep. 2015 May;42(5):963-75. doi: 10.1007/s11033-014-3833-y. Epub 2014 Nov 16. Mol Biol Rep. 2015. PMID: 25399080
-
The glutaminase (CgGLS-1) mediates anti-bacterial immunity by prompting cytokine synthesis and hemocyte apoptosis in Pacific oyster Crassostrea gigas.Sci Rep. 2021 Jan 14;11(1):1281. doi: 10.1038/s41598-020-80552-2. Sci Rep. 2021. PMID: 33446806 Free PMC article.
-
Sublethal exposure of eastern oyster Crassostrea virginica to the goniodomin-producing dinoflagellate Alexandrium monilatum: Fate of toxins, histopathology, and gene expression.J Aquat Anim Health. 2024 Dec;36(4):374-394. doi: 10.1002/aah.10227. J Aquat Anim Health. 2024. PMID: 39739761 Free PMC article.
-
In vitro Evaluation of Programmed Cell Death in the Immune System of Pacific Oyster Crassostrea gigas by the Effect of Marine Toxins.Front Immunol. 2021 Apr 1;12:634497. doi: 10.3389/fimmu.2021.634497. eCollection 2021. Front Immunol. 2021. PMID: 33868255 Free PMC article.
-
The polymorphism in the promoter of HSP70 gene is associated with heat tolerance of two congener endemic bay scallops (Argopecten irradians irradians and A. i. concentricus).PLoS One. 2014 Jul 16;9(7):e102332. doi: 10.1371/journal.pone.0102332. eCollection 2014. PLoS One. 2014. PMID: 25028964 Free PMC article.
References
-
- Hacker G (2000) The morphology of apoptosis. Cell Tissue Res 301: 5–17. - PubMed
-
- Kang TB, Ben-Moshe T, Varfolomeev EE, Pewzner-Jung Y, Yogev N, et al. (2004) Caspase-8 serves both apoptotic and nonapoptotic roles. Journal of Immunology 173: 2976–2984. - PubMed
-
- Nicholson DW (1999) Caspase structure, proteolytic substrates, and function during apoptotic cell death. Cell Death Differ 6: 1028–1042. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

