Binding of histone H1 to DNA is differentially modulated by redox state of HMGB1
- PMID: 24551219
- PMCID: PMC3923860
- DOI: 10.1371/journal.pone.0089070
Binding of histone H1 to DNA is differentially modulated by redox state of HMGB1
Abstract
HMGB1 is an architectural protein in chromatin, acting also as a signaling molecule outside the cell. Recent reports from several laboratories provided evidence that a number of both the intracellular and extracellular functions of HMGB1 may depend on redox-sensitive cysteine residues of the protein. In this study we demonstrate that redox state of HMGB1 can significantly modulate the ability of the protein to bind and bend DNA, as well as to promote DNA end-joining. We also report a high affinity binding of histone H1 to hemicatenated DNA loops and DNA minicircles. Finally, we show that reduced HMGB1 can readily displace histone H1 from DNA, while oxidized HMGB1 has limited capacity for H1 displacement. Our results suggested a novel mechanism for the HMGB1-mediated modulation of histone H1 binding to DNA. Possible biological consequences of linker histones H1 replacement by HMGB1 for the functioning of chromatin are discussed.
Conflict of interest statement
Figures
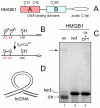
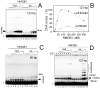



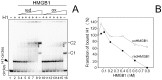
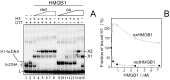
References
-
- Štros M (2010) HMGB proteins: interactions with DNA and chromatin. Biochim Biophys Acta 1799: 101–113. - PubMed
-
- Calogero S, Grassi F, Aguzzi A, Voigtlander T, Ferrier P, et al. (1999) The lack of chromosomal protein Hmg1 does not disrupt cell growth but causes lethal hypoglycaemia in newborn mice. Nat Genet 22: 276–280. - PubMed
-
- Hoppe G, Talcott KE, Bhattacharya SK, Crabb JW, Sears JE (2006) Molecular basis for the redox control of nuclear transport of the structural chromatin protein Hmgb1. Exp Cell Res 312: 3526–3538. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

