Potential of cometabolic transformation of polysaccharides and lignin in lignocellulose by soil Actinobacteria
- PMID: 24551229
- PMCID: PMC3923840
- DOI: 10.1371/journal.pone.0089108
Potential of cometabolic transformation of polysaccharides and lignin in lignocellulose by soil Actinobacteria
Abstract
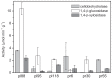
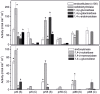
While it is known that several Actinobacteria produce enzymes that decompose polysaccharides or phenolic compounds in dead plant biomass, the occurrence of these traits in the environment remains largely unclear. The aim of this work was to screen isolated actinobacterial strains to explore their ability to produce extracellular enzymes that participate in the degradation of polysaccharides and their ability to cometabolically transform phenolic compounds of various complexities. Actinobacterial strains were isolated from meadow and forest soils and screened for their ability to grow on lignocellulose. The potential to transform (14)C-labelled phenolic substrates (dehydrogenation polymer (DHP), lignin and catechol) and to produce a range of extracellular, hydrolytic enzymes was investigated in three strains of Streptomyces spp. that possessed high lignocellulose degrading activity. Isolated strains showed high variation in their ability to produce cellulose- and hemicellulose-degrading enzymes and were able to mineralise up to 1.1% and to solubilise up to 4% of poplar lignin and to mineralise up to 11.4% and to solubilise up to 64% of catechol, while only minimal mineralisation of DHP was observed. The results confirm the potential importance of Actinobacteria in lignocellulose degradation, although it is likely that the decomposition of biopolymers is limited to strains that represent only a minor portion of the entire community, while the range of simple, carbon-containing compounds that serve as sources for actinobacterial growth is relatively wide.
Conflict of interest statement
Figures

Similar articles
-
Cellulolytic Streptomyces strains associated with herbivorous insects share a phylogenetically linked capacity to degrade lignocellulose.Appl Environ Microbiol. 2014 Aug;80(15):4692-701. doi: 10.1128/AEM.01133-14. Appl Environ Microbiol. 2014. PMID: 24837391 Free PMC article.
-
Use of lignocellulose biomass for endoxylanase production by Streptomyces termitum.Prep Biochem Biotechnol. 2017 May 28;47(5):505-512. doi: 10.1080/10826068.2016.1275015. Epub 2017 Jan 3. Prep Biochem Biotechnol. 2017. PMID: 28045607
-
Potential of semiarid soil from Caatinga biome as a novel source for mining lignocellulose-degrading enzymes.FEMS Microbiol Ecol. 2017 Feb;93(2):fiw248. doi: 10.1093/femsec/fiw248. Epub 2016 Dec 15. FEMS Microbiol Ecol. 2017. PMID: 27986827
-
Biodegradation and biological treatments of cellulose, hemicellulose and lignin: an overview.Int Microbiol. 2002 Jun;5(2):53-63. doi: 10.1007/s10123-002-0062-3. Int Microbiol. 2002. PMID: 12180781 Review.
-
Proteomic researches for lignocellulose-degrading enzymes: A mini-review.Bioresour Technol. 2018 Oct;265:532-541. doi: 10.1016/j.biortech.2018.05.101. Epub 2018 May 31. Bioresour Technol. 2018. PMID: 29884341 Review.
Cited by
-
Bacteria from the endosphere and rhizosphere of Quercus spp. use mainly cell wall-associated enzymes to decompose organic matter.PLoS One. 2019 Mar 25;14(3):e0214422. doi: 10.1371/journal.pone.0214422. eCollection 2019. PLoS One. 2019. PMID: 30908541 Free PMC article.
-
Interruption after Short-Term Nitrogen Additions Improves Ecological Stability of Larix olgensis Forest Soil by Affecting Bacterial Communities.Microorganisms. 2024 May 11;12(5):969. doi: 10.3390/microorganisms12050969. Microorganisms. 2024. PMID: 38792798 Free PMC article.
-
Nutrient availability is a dominant predictor of soil bacterial and fungal community composition after nitrogen addition in subtropical acidic forests.PLoS One. 2021 Feb 23;16(2):e0246263. doi: 10.1371/journal.pone.0246263. eCollection 2021. PLoS One. 2021. PMID: 33621258 Free PMC article.
-
Microbial community diversity and assembly processes in the aridification of wetlands on the Qinghai-Tibet Plateau.iScience. 2025 Apr 21;28(5):112494. doi: 10.1016/j.isci.2025.112494. eCollection 2025 May 16. iScience. 2025. PMID: 40395670 Free PMC article.
-
Response of Rhizosphere Bacterial Communities to Near-Natural Forest Management and Tree Species within Chinese Fir Plantations.Microbiol Spectr. 2023 Feb 14;11(1):e0232822. doi: 10.1128/spectrum.02328-22. Epub 2023 Jan 23. Microbiol Spectr. 2023. PMID: 36688690 Free PMC article.
References
-
- Baldrian P, Šnajdr J (2011) Lignocellulose-Degrading Enzymes in Soils. In: Shukla G, Varma A, editors. Soil Enzymology. Berlin: Springer-Verlag Berlin. p.167–186.
-
- de Boer W, Folman LB, Summerbell RC, Boddy L (2005) Living in a fungal world: impact of fungi on soil bacterial niche development. FEMS Microbiol Rev 29: 795–811. - PubMed
-
- Baldrian P (2008) Enzymes of Saprotrophic Basidiomycetes. In: Boddy L, Frankland JC, VanWest P, editors. Ecology of Saprotrophic Basidiomycetes. San Diego: Elsevier Academic Press Inc. p.19–41.
-
- Eastwood DC, Floudas D, Binder M, Majcherczyk A, Schneider P, et al. (2011) The Plant Cell Wall–Decomposing Machinery Underlies the Functional Diversity of Forest Fungi. Science 333: 762–765. - PubMed
-
- Floudas D, Binder M, Riley R, Barry K, Blanchette RA, et al. (2012) The paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science 336: 1715–1719. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

