Inferring the semantic relationships of words within an ontology using random indexing: applications to pharmacogenomics
- PMID: 24551397
- PMCID: PMC3900134
Inferring the semantic relationships of words within an ontology using random indexing: applications to pharmacogenomics
Abstract
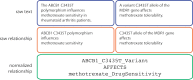
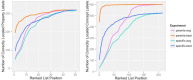
The biomedical literature presents a uniquely challenging text mining problem. Sentences are long and complex, the subject matter is highly specialized with a distinct vocabulary, and producing annotated training data for this domain is time consuming and expensive. In this environment, unsupervised text mining methods that do not rely on annotated training data are valuable. Here we investigate the use of random indexing, an automated method for producing vector-space semantic representations of words from large, unlabeled corpora, to address the problem of term normalization in sentences describing drugs and genes. We show that random indexing produces similarity scores that capture some of the structure of PHARE, a manually curated ontology of pharmacogenomics concepts. We further show that random indexing can be used to identify likely word candidates for inclusion in the ontology, and can help localize these new labels among classes and roles within the ontology.
Figures
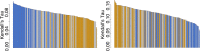
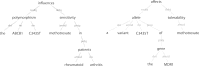
References
-
- Turney PD, Pantel P. From frequency to meaning: Vector space models of semantics. Journal of Artificial Intelligence Research. 2010;37(1):141–188.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
