Intragenic homogenization and multiple copies of prey-wrapping silk genes in Argiope garden spiders
- PMID: 24552485
- PMCID: PMC3933166
- DOI: 10.1186/1471-2148-14-31
Intragenic homogenization and multiple copies of prey-wrapping silk genes in Argiope garden spiders
Abstract
Background: Spider silks are spectacular examples of phenotypic diversity arising from adaptive molecular evolution. An individual spider can produce an array of specialized silks, with the majority of constituent silk proteins encoded by members of the spidroin gene family. Spidroins are dominated by tandem repeats flanked by short, non-repetitive N- and C-terminal coding regions. The remarkable mechanical properties of spider silks have been largely attributed to the repeat sequences. However, the molecular evolutionary processes acting on spidroin terminal and repetitive regions remain unclear due to a paucity of complete gene sequences and sampling of genetic variation among individuals. To better understand spider silk evolution, we characterize a complete aciniform spidroin gene from an Argiope orb-weaving spider and survey aciniform gene fragments from congeneric individuals.
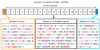
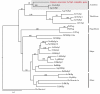
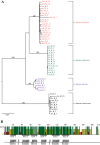
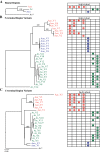
Results: We present the complete aciniform spidroin (AcSp1) gene from the silver garden spider Argiope argentata (Aar_AcSp1), and document multiple AcSp1 loci in individual genomes of A. argentata and the congeneric A. trifasciata and A. aurantia. We find that Aar_AcSp1 repeats have >98% pairwise nucleotide identity. By comparing AcSp1 repeat amino acid sequences between Argiope species and with other genera, we identify regions of conservation over vast amounts of evolutionary time. Through a PCR survey of individual A. argentata, A. trifasciata, and A. aurantia genomes, we ascertain that AcSp1 repeats show limited variation between species whereas terminal regions are more divergent. We also find that average dN/dS across codons in the N-terminal, repetitive, and C-terminal encoding regions indicate purifying selection that is strongest in the N-terminal region.
Conclusions: Using the complete A. argentata AcSp1 gene and spidroin genetic variation between individuals, this study clarifies some of the molecular evolutionary processes underlying the spectacular mechanical attributes of aciniform silk. It is likely that intragenic concerted evolution and functional constraints on A. argentata AcSp1 repeats result in extreme repeat homogeneity. The maintenance of multiple AcSp1 encoding loci in Argiope genomes supports the hypothesis that Argiope spiders require rapid and efficient protein production to support their prolific use of aciniform silk for prey-wrapping and web-decorating. In addition, multiple gene copies may represent the early stages of spidroin diversification.
Figures
References
-
- Vasanthavada K, Hu X, Falick A, Mattina CL, Moore A, Jones P, Yee R, Reza R, Tuton T, Vierra C. Aciniform spidroin, a constituent of egg case sacs and wrapping silk fibers from the black widow spider Latrodectus hesperus. J Biol Chem. 2007;282:35088–35097. doi: 10.1074/jbc.M705791200. - DOI - PubMed
-
- Griswold CE, Coddington JA, Hormiga G, Scharff N. Phylogeny of the orb-web building spiders (Araneae, Orbiculariae: Deinopoidea, Araneoidea) Zool J Linn Soc-Lond. 1998;123:1–99. doi: 10.1111/j.1096-3642.1998.tb01290.x. - DOI
-
- Walter A, Elgar MA, Bliss P, Moritz RFA. Wrap attack activates web-decorating behavior in Argiope spiders. Behav Ecol. 2008;19:799–804. doi: 10.1093/beheco/arn030. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

