The dharma of nonsense-mediated mRNA decay in mammalian cells
- PMID: 24552703
- PMCID: PMC3907001
- DOI: 10.14348/molcells.2014.2193
The dharma of nonsense-mediated mRNA decay in mammalian cells
Abstract
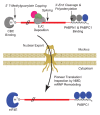
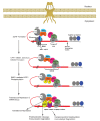
Mammalian-cell messenger RNAs (mRNAs) are generated in the nucleus from precursor RNAs (pre-mRNAs, which often contain one or more introns) that are complexed with an array of incompletely inventoried proteins. During their biogenesis, pre-mRNAs and their derivative mRNAs are subject to extensive cis-modifications. These modifications promote the binding of distinct polypeptides that mediate a diverse array of functions needed for mRNA metabolism, including nuclear export, inspection by the nonsense-mediated mRNA decay (NMD) quality-control machinery, and synthesis of the encoded protein product. Ribonucleoprotein complex (RNP) remodeling through the loss and gain of protein constituents before and after pre-mRNA splicing, during mRNA export, and within the cytoplasm facilitates NMD, ensuring integrity of the transcriptome. Here we review the mRNP rearrangements that culminate in detection and elimination of faulty transcripts by mammalian-cell NMD.
Figures
References
-
- Amrani N., Ganesan R., Kervestin S., Mangus D.A., Ghosh S., Jacobson A. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature. 2004;432:112–118. - PubMed
-
- Anczukow O., Ware M.D., Buisson M., Zetoune A.B., Stoppa-Lyonnet D., Sinilnikova O.M., Mazoyer S. Does the nonsense-mediated mRNA decay mechanism prevent the synthesis of truncated BRCA1, CHK2, and p53 proteins? Hum. Mutat. 2008;29:65–73. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

