Listeria monocytogenes induces host DNA damage and delays the host cell cycle to promote infection
- PMID: 24552813
- PMCID: PMC3984316
- DOI: 10.4161/cc.27780
Listeria monocytogenes induces host DNA damage and delays the host cell cycle to promote infection
Abstract
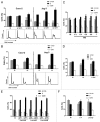
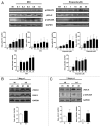
Listeria monocytogenes (Lm) is a human intracellular pathogen widely used to uncover the mechanisms evolved by pathogens to establish infection. However, its capacity to perturb the host cell cycle was never reported. We show that Lm infection affects the host cell cycle progression, increasing its overall duration but allowing consecutive rounds of division. A complete Lm infectious cycle induces a S-phase delay accompanied by a slower rate of DNA synthesis and increased levels of host DNA strand breaks. Additionally, DNA damage/replication checkpoint responses are triggered in an Lm dose-dependent manner through the phosphorylation of DNA-PK, H2A.X, and CDC25A and independently from ATM/ATR. While host DNA damage induced exogenously favors Lm dissemination, the override of checkpoint pathways limits infection. We propose that host DNA replication disturbed by Lm infection culminates in DNA strand breaks, triggering DNA damage/replication responses, and ensuring a cell cycle delay that favors Lm propagation.
Keywords: CDC25A; DNA damage; DNA-PK; Listeria monocytogenes; bacterial infection; host cell cycle.
Figures





Similar articles
-
Listeria monocytogenes Interferes with Host Cell Mitosis through Its Virulence Factors InlC and ActA.Toxins (Basel). 2020 Jun 20;12(6):411. doi: 10.3390/toxins12060411. Toxins (Basel). 2020. PMID: 32575670 Free PMC article.
-
Lmo1656 is a secreted virulence factor of Listeria monocytogenes that interacts with the sorting nexin 6-BAR complex.J Biol Chem. 2018 Jun 15;293(24):9265-9276. doi: 10.1074/jbc.RA117.000365. Epub 2018 Apr 17. J Biol Chem. 2018. PMID: 29666193 Free PMC article.
-
Type I interferon promotes cell-to-cell spread of Listeria monocytogenes.Cell Microbiol. 2017 Mar;19(3). doi: 10.1111/cmi.12660. Epub 2016 Oct 10. Cell Microbiol. 2017. PMID: 27582004
-
In vitro and in vivo models to study human listeriosis: mind the gap.Microbes Infect. 2013 Dec;15(14-15):971-80. doi: 10.1016/j.micinf.2013.09.012. Epub 2013 Oct 18. Microbes Infect. 2013. PMID: 24144539 Review.
-
The bacterial pathogen Listeria monocytogenes and the interferon family: type I, type II and type III interferons.Front Cell Infect Microbiol. 2014 Apr 28;4:50. doi: 10.3389/fcimb.2014.00050. eCollection 2014. Front Cell Infect Microbiol. 2014. PMID: 24809023 Free PMC article. Review.
Cited by
-
Tampering of Viruses and Bacteria with Host DNA Repair: Implications for Cellular Transformation.Cancers (Basel). 2021 Jan 11;13(2):241. doi: 10.3390/cancers13020241. Cancers (Basel). 2021. PMID: 33440726 Free PMC article. Review.
-
Modulation of p53 during bacterial infections.Nat Rev Microbiol. 2015 Dec;13(12):741-8. doi: 10.1038/nrmicro3537. Epub 2015 Nov 9. Nat Rev Microbiol. 2015. PMID: 26548915 Review.
-
When our genome is targeted by pathogenic bacteria.Cell Mol Life Sci. 2015 Jul;72(14):2665-76. doi: 10.1007/s00018-015-1900-8. Epub 2015 Apr 17. Cell Mol Life Sci. 2015. PMID: 25877988 Free PMC article. Review.
-
Mycobacterium tuberculosis exploits host ATM kinase for survival advantage through SecA2 secretome.Elife. 2020 Mar 30;9:e51466. doi: 10.7554/eLife.51466. Elife. 2020. PMID: 32223892 Free PMC article.
-
Listeria monocytogenes Interferes with Host Cell Mitosis through Its Virulence Factors InlC and ActA.Toxins (Basel). 2020 Jun 20;12(6):411. doi: 10.3390/toxins12060411. Toxins (Basel). 2020. PMID: 32575670 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
