Generation of mastitis resistance in cows by targeting human lysozyme gene to β-casein locus using zinc-finger nucleases
- PMID: 24552841
- PMCID: PMC4027401
- DOI: 10.1098/rspb.2013.3368
Generation of mastitis resistance in cows by targeting human lysozyme gene to β-casein locus using zinc-finger nucleases
Abstract
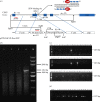
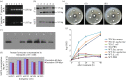
Mastitis costs the dairy industry billions of dollars annually and is the most consequential disease of dairy cattle. Transgenic cows secreting an antimicrobial peptide demonstrated resistance to mastitis. The combination of somatic cell gene targeting and nuclear transfer provides a powerful method to produce transgenic animals. Recent studies found that a precisely placed double-strand break induced by engineered zinc-finger nucleases (ZFNs) stimulated the integration of exogenous DNA stretches into a pre-determined genomic location, resulting in high-efficiency site-specific gene addition. Here, we used ZFNs to target human lysozyme (hLYZ) gene to bovine β-casein locus, resulting in hLYZ knock-in of approximately 1% of ZFN-treated bovine fetal fibroblasts (BFFs). Gene-targeted fibroblast cell clones were screened by junction PCR amplification and Southern blot analysis. Gene-targeted BFFs were used in somatic cell nuclear transfer. In vitro assays demonstrated that the milk secreted by transgenic cows had the ability to kill Staphylococcus aureus. We report the production of cloned cows carrying human lysozyme gene knock-in β-casein locus using ZFNs. Our findings open a unique avenue for the creation of transgenic cows from genetic engineering by providing a viable tool for enhancing resistance to disease and improving the health and welfare of livestock.
Keywords: gene targeting; mastitis; nuclear transfer; zinc-finger nucleases; β-casein locus.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
