Stalled transcription complexes promote DNA repair at a distance
- PMID: 24554077
- PMCID: PMC3964087
- DOI: 10.1073/pnas.1322350111
Stalled transcription complexes promote DNA repair at a distance
Abstract
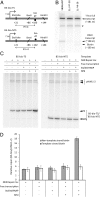
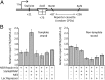
Transcription-coupled nucleotide excision repair (TCR) accelerates the removal of noncoding lesions from the template strand of active genes, and hence contributes to genome-wide variations in mutation frequency. Current models for TCR suppose that a lesion must cause RNA polymerase (RNAP) to stall if it is to be a substrate for accelerated repair. We have examined the substrate requirements for TCR using a system in which transcription stalling and damage location can be uncoupled. We show that Mfd-dependent TCR in bacteria involves the formation of a damage search complex that can detect lesions downstream of a stalled RNAP, and that the strand specificity of the accelerated repair pathway is independent of the requirement for a lesion to stall RNAP. We also show that an ops (operon polarity suppressor) transcription pause site, which causes backtracking of RNAP, can promote the repair of downstream lesions when those lesions do not themselves cause the polymerase to stall. Our findings indicate that the transcription-repair coupling factor Mfd, which is an ATP-dependent superfamily 2 helicase that binds to RNAP, continues to translocate along DNA after RNAP has been displaced until a lesion in the template strand is located. The discovery that pause sites can promote the repair of nonstalling lesions suggests that TCR pathways may play a wider role in modulating mutation frequencies in different parts of the genome than has previously been suspected.
Keywords: ATPase; DNA translocase; UvrA; protein roadblock.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Comment in
-
Transcriptional pausing to scout ahead for DNA damage.Proc Natl Acad Sci U S A. 2014 Mar 18;111(11):3905-6. doi: 10.1073/pnas.1402020111. Epub 2014 Mar 5. Proc Natl Acad Sci U S A. 2014. PMID: 24599593 Free PMC article. No abstract available.
References
-
- Martincorena I, Seshasayee ASN, Luscombe NM. Evidence of non-random mutation rates suggests an evolutionary risk management strategy. Nature. 2012;485(7396):95–98. - PubMed
-
- Hanawalt PC, Spivak G. Transcription-coupled DNA repair: Two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9(12):958–970. - PubMed
-
- Mellon I, Hanawalt PC. Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed DNA strand. Nature. 1989;342(6245):95–98. - PubMed
-
- Martincorena I, Luscombe NM. Non-random mutation: The evolution of targeted hypermutation and hypomutation. Bioessays. 2013;35(2):123–130. - PubMed
-
- Borukhov S, Nudler E. RNA polymerase: The vehicle of transcription. Trends Microbiol. 2008;16(3):126–134. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

