Dicer-like 3 produces transposable element-associated 24-nt siRNAs that control agricultural traits in rice
- PMID: 24554078
- PMCID: PMC3956178
- DOI: 10.1073/pnas.1318131111
Dicer-like 3 produces transposable element-associated 24-nt siRNAs that control agricultural traits in rice
Abstract
Transposable elements (TEs) and repetitive sequences make up over 35% of the rice (Oryza sativa) genome. The host regulates the activity of different TEs by different epigenetic mechanisms, including DNA methylation, histone H3K9 methylation, and histone H3K4 demethylation. TEs can also affect the expression of host genes. For example, miniature inverted repeat TEs (MITEs), dispersed high copy-number DNA TEs, can influence the expression of nearby genes. In plants, 24-nt small interfering RNAs (siRNAs) are mainly derived from repeats and TEs. However, the extent to which TEs, particularly MITEs associated with 24-nt siRNAs, affect gene expression remains elusive. Here, we show that the rice Dicer-like 3 homolog OsDCL3a is primarily responsible for 24-nt siRNA processing. Impairing OsDCL3a expression by RNA interference caused phenotypes affecting important agricultural traits; these phenotypes include dwarfism, larger flag leaf angle, and fewer secondary branches. We used small RNA deep sequencing to identify 535,054 24-nt siRNA clusters. Of these clusters, ∼82% were OsDCL3a-dependent and showed significant enrichment of MITEs. Reduction of OsDCL3a function reduced the 24-nt siRNAs predominantly from MITEs and elevated expression of nearby genes. OsDCL3a directly targets genes involved in gibberellin and brassinosteroid homeostasis; OsDCL3a deficiency may affect these genes, thus causing the phenotypes of dwarfism and enlarged flag leaf angle. Our work identifies OsDCL3a-dependent 24-nt siRNAs derived from MITEs as broadly functioning regulators for fine-tuning gene expression, which may reflect a conserved epigenetic mechanism in higher plants with genomes rich in dispersed repeats or TEs.
Keywords: plant architecture; plant hormone; transposon.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

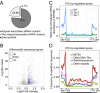


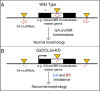
Similar articles
-
Regulation of Rice Tillering by RNA-Directed DNA Methylation at Miniature Inverted-Repeat Transposable Elements.Mol Plant. 2020 Jun 1;13(6):851-863. doi: 10.1016/j.molp.2020.02.009. Epub 2020 Feb 19. Mol Plant. 2020. PMID: 32087371
-
Identification of miniature inverted-repeat transposable elements (MITEs) and biogenesis of their siRNAs in the Solanaceae: new functional implications for MITEs.Genome Res. 2009 Jan;19(1):42-56. doi: 10.1101/gr.078196.108. Epub 2008 Nov 26. Genome Res. 2009. PMID: 19037014 Free PMC article.
-
Role of transposon-derived small RNAs in the interplay between genomes and parasitic DNA in rice.PLoS Genet. 2012 Sep;8(9):e1002953. doi: 10.1371/journal.pgen.1002953. Epub 2012 Sep 27. PLoS Genet. 2012. PMID: 23028360 Free PMC article.
-
Miniature inverted-repeat transposable elements (MITEs), derived insertional polymorphism as a tool of marker systems for molecular plant breeding.Mol Biol Rep. 2020 Apr;47(4):3155-3167. doi: 10.1007/s11033-020-05365-y. Epub 2020 Mar 11. Mol Biol Rep. 2020. PMID: 32162128 Review.
-
Small RNAs and transposon silencing in plants.Dev Growth Differ. 2012 Jan;54(1):100-7. doi: 10.1111/j.1440-169X.2011.01309.x. Epub 2011 Dec 12. Dev Growth Differ. 2012. PMID: 22150226 Review.
Cited by
-
Non-coding RNA polymerases that silence transposable elements and reprogram gene expression in plants.Transcription. 2020 Jun-Aug;11(3-4):172-191. doi: 10.1080/21541264.2020.1825906. Epub 2020 Nov 12. Transcription. 2020. PMID: 33180661 Free PMC article. Review.
-
Phytohormone-Mediated Molecular Mechanisms Involving Multiple Genes and QTL Govern Grain Number in Rice.Front Genet. 2020 Nov 12;11:586462. doi: 10.3389/fgene.2020.586462. eCollection 2020. Front Genet. 2020. PMID: 33281879 Free PMC article. Review.
-
DREAM complex suppresses DNA methylation maintenance genes and precludes DNA hypermethylation.Nat Plants. 2020 Aug;6(8):942-956. doi: 10.1038/s41477-020-0710-7. Epub 2020 Jul 13. Nat Plants. 2020. PMID: 32661276
-
Stowaway miniature inverted repeat transposable elements are important agents driving recent genomic diversity in wild and cultivated carrot.Mob DNA. 2019 Nov 27;10:47. doi: 10.1186/s13100-019-0190-3. eCollection 2019. Mob DNA. 2019. PMID: 31798695 Free PMC article.
-
Methylation of a MITE insertion in the MdRFNR1-1 promoter is positively associated with its allelic expression in apple in response to drought stress.Plant Cell. 2022 Sep 27;34(10):3983-4006. doi: 10.1093/plcell/koac220. Plant Cell. 2022. PMID: 35897144 Free PMC article.
References
-
- International Rice Genome Sequencing Project The map-based sequence of the rice genome. Nature. 2005;436(7052):793–800. - PubMed
-
- Feschotte C, Jiang N, Wessler SR. Plant transposable elements: Where genetics meets genomics. Nat Rev Genet. 2002;3(5):329–341. - PubMed
-
- Lisch D. How important are transposons for plant evolution? Nat Rev Genet. 2013;14(1):49–61. - PubMed
-
- Jiang N, Feschotte C, Zhang X, Wessler SR. Using rice to understand the origin and amplification of miniature inverted repeat transposable elements (MITEs) Curr Opin Plant Biol. 2004;7(2):115–119. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

