The life cycle of Drosophila orphan genes
- PMID: 24554240
- PMCID: PMC3927632
- DOI: 10.7554/eLife.01311
The life cycle of Drosophila orphan genes
Abstract
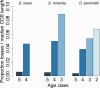
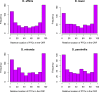
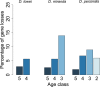
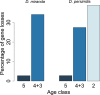
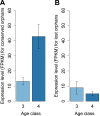
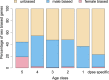
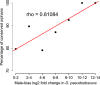
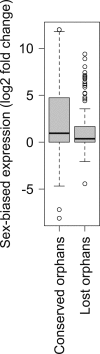
Orphans are genes restricted to a single phylogenetic lineage and emerge at high rates. While this predicts an accumulation of genes, the gene number has remained remarkably constant through evolution. This paradox has not yet been resolved. Because orphan genes have been mainly analyzed over long evolutionary time scales, orphan loss has remained unexplored. Here we study the patterns of orphan turnover among close relatives in the Drosophila obscura group. We show that orphans are not only emerging at a high rate, but that they are also rapidly lost. Interestingly, recently emerged orphans are more likely to be lost than older ones. Furthermore, highly expressed orphans with a strong male-bias are more likely to be retained. Since both lost and retained orphans show similar evolutionary signatures of functional conservation, we propose that orphan loss is not driven by high rates of sequence evolution, but reflects lineage-specific functional requirements. DOI: http://dx.doi.org/10.7554/eLife.01311.001.
Keywords: D. pseudoobscura; evolutionary rates; gene gains; gene losses; pseudogenes; sex-biased expression.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures










Comment in
-
Evolution: dynamics of de novo gene emergence.Curr Biol. 2014 Mar 17;24(6):R238-40. doi: 10.1016/j.cub.2014.02.016. Curr Biol. 2014. PMID: 24650912
References
-
- Beckenbach AT, Wei YW, Liu H. 1993. Relationships in the Drosophila obscura species group, inferred from mitochondrial cytochrome oxidase II sequences. Molecular Biology and Evolution 10:619–634 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

