Distinctive kinetics and substrate specificities of plant and fungal tRNA ligases
- PMID: 24554441
- PMCID: PMC3964908
- DOI: 10.1261/rna.043752.113
Distinctive kinetics and substrate specificities of plant and fungal tRNA ligases
Abstract
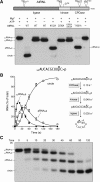
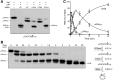
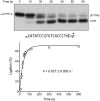
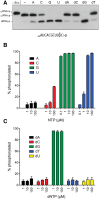
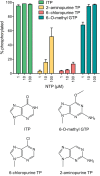
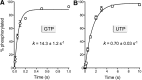
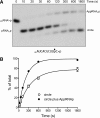
Plant and fungal tRNA ligases are trifunctional enzymes that repair RNA breaks with 2',3'-cyclic-PO4 and 5'-OH ends. They are composed of cyclic phosphodiesterase (CPDase) and polynucleotide kinase domains that heal the broken ends to generate the 3'-OH, 2'-PO4, and 5'-PO4 required for sealing by a ligase domain. Here, we use short HORNA>p substrates to determine, in a one-pot assay format under single-turnover conditions, the order and rates of the CPDase, kinase and ligase steps. The observed reaction sequence for the plant tRNA ligase AtRNL, independent of RNA length, is that the CPDase engages first, converting HORNA>p to HORNA2'p, which is then phosphorylated to pRNA2'p by the kinase. Whereas the rates of the AtRNL CPDase and kinase reactions are insensitive to RNA length, the rate of the ligase reaction is slowed by a factor of 16 in the transition from 10-mer RNA to 8-mer and further by eightfold in the transition from 8-mer RNA to 6-mer. We report that a single ribonucleoside-2',3'-cyclic-PO4 moiety enables AtRNL to efficiently splice an otherwise all-DNA strand. Our characterization of a fungal tRNA ligase (KlaTrl1) highlights important functional distinctions vis à vis the plant homolog. We find that (1) the KlaTrl1 kinase is 300-fold faster than the AtRNL kinase; and (2) the KlaTrl1 kinase is highly specific for GTP or dGTP as the phosphate donor. Our findings recommend tRNA ligase as a tool to map ribonucleotides embedded in DNA and as a target for antifungal drug discovery.
Keywords: 2′,3′ cyclic phosphodiesterase; RNA ligase; RNA repair; polynucleotide kinase.
Figures


Similar articles
-
Structure and two-metal mechanism of fungal tRNA ligase.Nucleic Acids Res. 2019 Feb 20;47(3):1428-1439. doi: 10.1093/nar/gky1275. Nucleic Acids Res. 2019. PMID: 30590734 Free PMC article.
-
Genetic and biochemical analysis of the functional domains of yeast tRNA ligase.J Biol Chem. 2003 Nov 7;278(45):43928-38. doi: 10.1074/jbc.M307839200. Epub 2003 Aug 21. J Biol Chem. 2003. PMID: 12933796
-
A kinetic framework for tRNA ligase and enforcement of a 2'-phosphate requirement for ligation highlights the design logic of an RNA repair machine.RNA. 2013 May;19(5):659-69. doi: 10.1261/rna.038406.113. Epub 2013 Mar 20. RNA. 2013. PMID: 23515942 Free PMC article.
-
RNA splicing in prokaryotes: bacteriophage T4 leads the way.Cell. 1985 Jun;41(2):339-40. doi: 10.1016/s0092-8674(85)80002-7. Cell. 1985. PMID: 2580641 Review. No abstract available.
-
RNA Repair: Hiding in Plain Sight.Annu Rev Genet. 2023 Nov 27;57:461-489. doi: 10.1146/annurev-genet-071719-021856. Epub 2023 Sep 18. Annu Rev Genet. 2023. PMID: 37722686 Review.
Cited by
-
Multiple decay events target HAC1 mRNA during splicing to regulate the unfolded protein response.Elife. 2019 Mar 15;8:e42262. doi: 10.7554/eLife.42262. Elife. 2019. PMID: 30874502 Free PMC article.
-
Structure and two-metal mechanism of fungal tRNA ligase.Nucleic Acids Res. 2019 Feb 20;47(3):1428-1439. doi: 10.1093/nar/gky1275. Nucleic Acids Res. 2019. PMID: 30590734 Free PMC article.
-
Substrate analogs that trap the 2'-phospho-ADP-ribosylated RNA intermediate of the Tpt1 (tRNA 2'-phosphotransferase) reaction pathway.RNA. 2020 Apr;26(4):373-381. doi: 10.1261/rna.074377.119. Epub 2020 Jan 13. RNA. 2020. PMID: 31932322 Free PMC article.
-
Structures of bacterial polynucleotide kinase in a michaelis complex with nucleoside triphosphate (NTP)-Mg2+ and 5'-OH RNA and a mixed substrate-product complex with NTP-Mg2+ and a 5'-phosphorylated oligonucleotide.J Bacteriol. 2014 Dec;196(24):4285-92. doi: 10.1128/JB.02197-14. Epub 2014 Sep 29. J Bacteriol. 2014. PMID: 25266383 Free PMC article.
-
Characterization of Runella slithyformis HD-Pnk, a Bifunctional DNA/RNA End-Healing Enzyme Composed of an N-Terminal 2',3'-Phosphoesterase HD Domain and a C-Terminal 5'-OH Polynucleotide Kinase Domain.J Bacteriol. 2017 Jan 12;199(3):e00739-16. doi: 10.1128/JB.00739-16. Print 2017 Feb 1. J Bacteriol. 2017. PMID: 27895092 Free PMC article.
References
-
- Apostol BL, Westaway SK, Abelson J, Greer CL 1991. Deletion analysis of a multifunctional yeast tRNA ligase polypeptide: Identification of essential and dispensable functional domains. J Biol Chem 266: 7445–7455 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
