Lymphocyte gene expression and JC virus noncoding control region sequences are linked with the risk of progressive multifocal leukoencephalopathy
- PMID: 24554653
- PMCID: PMC3993791
- DOI: 10.1128/JVI.03221-13
Lymphocyte gene expression and JC virus noncoding control region sequences are linked with the risk of progressive multifocal leukoencephalopathy
Abstract
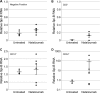
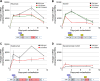
Progressive multifocal leukoencephalopathy (PML)-derived noncoding control region (NCCR) sequences permitted greater early viral gene expression than kidney-associated NCCR sequences. This was driven in part by binding of the transcription factor Spi-B to unique PML-associated Spi-B binding sites. Spi-B is upregulated in developing B cells in response to natalizumab therapy, a known risk factor for PML. Naturally occurring JCV sequence variation, together with drug treatment-induced cellular changes, may synergize to create an environment leading to an increased risk of PML.
Figures

Similar articles
-
Rearranged JC virus noncoding control regions found in progressive multifocal leukoencephalopathy patient samples increase virus early gene expression and replication rate.J Virol. 2010 Oct;84(20):10448-56. doi: 10.1128/JVI.00614-10. Epub 2010 Aug 4. J Virol. 2010. PMID: 20686041 Free PMC article.
-
Sequencing and analysis of JC virus DNA from natalizumab-treated PML patients.J Infect Dis. 2011 Jul 15;204(2):237-44. doi: 10.1093/infdis/jir256. J Infect Dis. 2011. PMID: 21673034 Free PMC article.
-
JC virus promoter/enhancers contain TATA box-associated Spi-B-binding sites that support early viral gene expression in primary astrocytes.J Gen Virol. 2012 Mar;93(Pt 3):651-661. doi: 10.1099/vir.0.035832-0. Epub 2011 Nov 9. J Gen Virol. 2012. PMID: 22071512 Free PMC article.
-
Polyomavirus JC in the context of immunosuppression: a series of adaptive, DNA replication-driven recombination events in the development of progressive multifocal leukoencephalopathy.Clin Dev Immunol. 2013;2013:197807. doi: 10.1155/2013/197807. Epub 2013 Apr 15. Clin Dev Immunol. 2013. PMID: 23690820 Free PMC article. Review.
-
[Genetic changes possibly associated with progressive multifocal leukoencephalopathy].Brain Nerve. 2007 Feb;59(2):109-18. Brain Nerve. 2007. PMID: 17380775 Review. Japanese.
Cited by
-
JC polyomavirus expression and bell-shaped regulation of its SF2/ASF suppressor during the follow-up of multiple sclerosis patients treated with natalizumab.J Neurovirol. 2017 Apr;23(2):226-238. doi: 10.1007/s13365-016-0492-x. Epub 2016 Nov 3. J Neurovirol. 2017. PMID: 27812788
-
Natalizumab Affects T-Cell Phenotype in Multiple Sclerosis: Implications for JCV Reactivation.PLoS One. 2016 Aug 3;11(8):e0160277. doi: 10.1371/journal.pone.0160277. eCollection 2016. PLoS One. 2016. PMID: 27486658 Free PMC article. Clinical Trial.
-
Progressive Multifocal Leukoencephalopathy in HIV-Uninfected Individuals.Curr Infect Dis Rep. 2016 Nov;18(11):33. doi: 10.1007/s11908-016-0543-8. Curr Infect Dis Rep. 2016. PMID: 27686675 Review.
-
JC Polyomavirus Abundance and Distribution in Progressive Multifocal Leukoencephalopathy (PML) Brain Tissue Implicates Myelin Sheath in Intracerebral Dissemination of Infection.PLoS One. 2016 May 18;11(5):e0155897. doi: 10.1371/journal.pone.0155897. eCollection 2016. PLoS One. 2016. PMID: 27191595 Free PMC article.
-
Natalizumab in Multiple Sclerosis Treatment: From Biological Effects to Immune Monitoring.Front Immunol. 2020 Sep 24;11:549842. doi: 10.3389/fimmu.2020.549842. eCollection 2020. Front Immunol. 2020. PMID: 33072089 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

