Epstein-Barr virus BALF3 has nuclease activity and mediates mature virion production during the lytic cycle
- PMID: 24554665
- PMCID: PMC3993834
- DOI: 10.1128/JVI.00063-14
Epstein-Barr virus BALF3 has nuclease activity and mediates mature virion production during the lytic cycle
Abstract
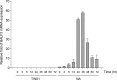
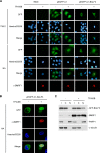
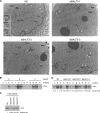
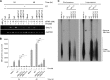
Epstein-Barr virus (EBV) lytic replication involves complex processes, including DNA synthesis, DNA cleavage and packaging, and virion egress. These processes require many different lytic gene products, but the mechanisms of their actions remain unclear, especially for DNA cleavage and packaging. According to sequence homology analysis, EBV BALF3, encoded by the third leftward open reading frame of the BamHI-A fragment in the viral genome, is a homologue of herpes simplex virus type 1 UL28. This gene product is believed to possess the properties of a terminase, such as nucleolytic activity on newly synthesized viral DNA and translocation of unit length viral genomes into procapsids. In order to characterize EBV BALF3, the protein was produced by and purified from recombinant baculoviruses and examined in an enzymatic reaction in vitro, which determined that EBV BALF3 acts as an endonuclease and its activity is modulated by Mg(2+), Mn(2+), and ATP. Moreover, in EBV-positive epithelial cells, BALF3 was expressed and transported from the cytoplasm into the nucleus following induction of the lytic cycle, and gene silencing of BALF3 caused a reduction of DNA packaging and virion release. Interestingly, suppression of BALF3 expression also decreased the efficiency of DNA synthesis. On the basis of these results, we suggest that EBV BALF3 is involved simultaneously in DNA synthesis and packaging and is required for the production of mature virions.
Importance: Virus lytic replication is essential to produce infectious virions, which is responsible for virus survival and spread. This work shows that an uncharacterized gene product of the human herpesvirus Epstein-Barr virus (EBV), BALF3, is expressed during the lytic cycle. In addition, BALF3 mediates an endonucleolytic reaction and is involved in viral DNA synthesis and packaging, leading to influence on the production of mature virions. According to sequence homology and physical properties, the lytic gene product BALF3 is considered a terminase in EBV. These findings identify a novel viral gene with an important role in contributing to a better understanding of the EBV life cycle.
Figures



References
-
- Nonoyama M, Tanaka A. 1975. Plasmid DNA as a possible state of Epstein-Barr virus genomes in nonproductive cells. Cold Spring Harbor Symp. Quant. Biol. 39:807–810 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

