The activating transcription factor 3 protein suppresses the oncogenic function of mutant p53 proteins
- PMID: 24554706
- PMCID: PMC3979409
- DOI: 10.1074/jbc.M113.503755
The activating transcription factor 3 protein suppresses the oncogenic function of mutant p53 proteins
Abstract
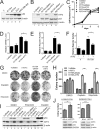
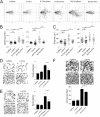
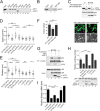
Mutant p53 proteins (mutp53) often acquire oncogenic activities, conferring drug resistance and/or promoting cancer cell migration and invasion. Although it has been well established that such a gain of function is mainly achieved through interaction with transcriptional regulators, thereby modulating cancer-associated gene expression, how the mutp53 function is regulated remains elusive. Here we report that activating transcription factor 3 (ATF3) bound common mutp53 (e.g. R175H and R273H) and, subsequently, suppressed their oncogenic activities. ATF3 repressed mutp53-induced NFKB2 expression and sensitized R175H-expressing cancer cells to cisplatin and etoposide treatments. Moreover, ATF3 appeared to suppress R175H- and R273H-mediated cancer cell migration and invasion as a consequence of preventing the transcription factor p63 from inactivation by mutp53. Accordingly, ATF3 promoted the expression of the metastasis suppressor SHARP1 in mutp53-expressing cells. An ATF3 mutant devoid of the mutp53-binding domain failed to disrupt the mutp53-p63 binding and, thus, lost the activity to suppress mutp53-mediated migration, suggesting that ATF3 binds to mutp53 to suppress its oncogenic function. In line with these results, we found that down-regulation of ATF3 expression correlated with lymph node metastasis in TP53-mutated human lung cancer. We conclude that ATF3 can suppress mutp53 oncogenic function, thereby contributing to tumor suppression in TP53-mutated cancer.
Keywords: ATF3; Drug Resistance; Invasion; Lung Cancer; Migration; Mutant p53; p53.
Figures




Similar articles
-
Plakoglobin restores tumor suppressor activity of p53R175H mutant by sequestering the oncogenic potential of β-catenin.Cancer Sci. 2018 Jun;109(6):1876-1888. doi: 10.1111/cas.13612. Epub 2018 May 23. Cancer Sci. 2018. PMID: 29660231 Free PMC article.
-
Gain-of-function mutant p53 upregulates CXC chemokines and enhances cell migration.Carcinogenesis. 2012 Feb;33(2):442-51. doi: 10.1093/carcin/bgr270. Epub 2011 Nov 22. Carcinogenesis. 2012. PMID: 22114072
-
SLC25A1, or CIC, is a novel transcriptional target of mutant p53 and a negative tumor prognostic marker.Oncotarget. 2014 Mar 15;5(5):1212-25. doi: 10.18632/oncotarget.1831. Oncotarget. 2014. PMID: 24681808 Free PMC article.
-
Mutant p53 in Cancer: Accumulation, Gain-of-Function, and Therapy.J Mol Biol. 2017 Jun 2;429(11):1595-1606. doi: 10.1016/j.jmb.2017.03.030. Epub 2017 Apr 6. J Mol Biol. 2017. PMID: 28390900 Free PMC article. Review.
-
p53 and its mutants in tumor cell migration and invasion.J Cell Biol. 2011 Jan 24;192(2):209-18. doi: 10.1083/jcb.201009059. J Cell Biol. 2011. PMID: 21263025 Free PMC article. Review.
Cited by
-
P53 Is Involved in a Three-Dimensional Architecture-Mediated Decrease in Chemosensitivity in Colon Cancer.J Cancer. 2016 Apr 29;7(8):900-9. doi: 10.7150/jca.14506. eCollection 2016. J Cancer. 2016. PMID: 27313779 Free PMC article.
-
P53 mutations in colorectal cancer - molecular pathogenesis and pharmacological reactivation.World J Gastroenterol. 2015 Jan 7;21(1):84-93. doi: 10.3748/wjg.v21.i1.84. World J Gastroenterol. 2015. PMID: 25574081 Free PMC article. Review.
-
From regulation to deregulation of p53 in hematologic malignancies: implications for diagnosis, prognosis and therapy.Biomark Res. 2024 Nov 14;12(1):137. doi: 10.1186/s40364-024-00676-9. Biomark Res. 2024. PMID: 39538363 Free PMC article. Review.
-
Decreased expression of ATF3, orchestrated by β-catenin/TCF3, miR-17-5p and HOXA11-AS, promoted gastric cancer progression via increased β-catenin and CEMIP.Exp Mol Med. 2021 Nov;53(11):1706-1722. doi: 10.1038/s12276-021-00694-9. Epub 2021 Nov 2. Exp Mol Med. 2021. PMID: 34728784 Free PMC article.
-
The common stress responsive transcription factor ATF3 binds genomic sites enriched with p300 and H3K27ac for transcriptional regulation.BMC Genomics. 2016 May 4;17:335. doi: 10.1186/s12864-016-2664-8. BMC Genomics. 2016. PMID: 27146783 Free PMC article.
References
-
- Brosh R, Rotter V. (2009) When mutants gain new powers. News from the mutant p53 field. Nat. Rev. Cancer 9, 701–713 - PubMed
-
- Olive K. P., Tuveson D. A., Ruhe Z. C., Yin B., Willis N. A., Bronson R. T., Crowley D., Jacks T. (2004) Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell 119, 847–860 - PubMed
-
- Lang G. A., Iwakuma T., Suh Y.-A., Liu G., Rao V. A., Parant J. M., Valentin-Vega Y. A., Terzian T., Caldwell L. C., Strong L. C., El-Naggar A. K., Lozano G. (2004) Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell 119, 861–872 - PubMed
-
- Muller P. A., Vousden K. H. (2013) p53 mutations in cancer. Nat. Cell Biol. 15, 2–8 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

