A genome-wide MeSH-based literature mining system predicts implicit gene-to-gene relationships and networks
- PMID: 24555475
- PMCID: PMC3852244
- DOI: 10.1186/1752-0509-7-S3-S9
A genome-wide MeSH-based literature mining system predicts implicit gene-to-gene relationships and networks
Abstract
Background: The large amount of literature in the post-genomics era enables the study of gene interactions and networks using all available articles published for a specific organism. MeSH is a controlled vocabulary of medical and scientific terms that is used by biomedical scientists to manually index articles in the PubMed literature database. We hypothesized that genome-wide gene-MeSH term associations from the PubMed literature database could be used to predict implicit gene-to-gene relationships and networks. While the gene-MeSH associations have been used to detect gene-gene interactions in some studies, different methods have not been well compared, and such a strategy has not been evaluated for a genome-wide literature analysis. Genome-wide literature mining of gene-to-gene interactions allows ranking of the best gene interactions and investigation of comprehensive biological networks at a genome level.
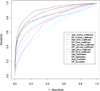
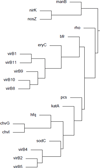
Results: The genome-wide GenoMesh literature mining algorithm was developed by sequentially generating a gene-article matrix, a normalized gene-MeSH term matrix, and a gene-gene matrix. The gene-gene matrix relies on the calculation of pairwise gene dissimilarities based on gene-MeSH relationships. An optimized dissimilarity score was identified from six well-studied functions based on a receiver operating characteristic (ROC) analysis. Based on the studies with well-studied Escherichia coli and less-studied Brucella spp., GenoMesh was found to accurately identify gene functions using weighted MeSH terms, predict gene-gene interactions not reported in the literature, and cluster all the genes studied from an organism using the MeSH-based gene-gene matrix. A web-based GenoMesh literature mining program is also available at: http://genomesh.hegroup.org. GenoMesh also predicts gene interactions and networks among genes associated with specific MeSH terms or user-selected gene lists.
Conclusions: The GenoMesh algorithm and web program provide the first genome-wide, MeSH-based literature mining system that effectively predicts implicit gene-gene interaction relationships and networks in a genome-wide scope.
Figures




Similar articles
-
Ontology-based Brucella vaccine literature indexing and systematic analysis of gene-vaccine association network.BMC Immunol. 2011 Aug 26;12:49. doi: 10.1186/1471-2172-12-49. BMC Immunol. 2011. PMID: 21871085 Free PMC article.
-
Ontology-based literature mining of E. coli vaccine-associated gene interaction networks.J Biomed Semantics. 2017 Mar 14;8(1):12. doi: 10.1186/s13326-017-0122-4. J Biomed Semantics. 2017. PMID: 28288685 Free PMC article.
-
BBP: Brucella genome annotation with literature mining and curation.BMC Bioinformatics. 2006 Jul 16;7:347. doi: 10.1186/1471-2105-7-347. BMC Bioinformatics. 2006. PMID: 16842628 Free PMC article.
-
Mining emerging biomedical literature for understanding disease associations in drug discovery.Methods Mol Biol. 2014;1159:171-206. doi: 10.1007/978-1-4939-0709-0_11. Methods Mol Biol. 2014. PMID: 24788268 Review.
-
Integrative literature and data mining to rank disease candidate genes.Methods Mol Biol. 2014;1159:207-26. doi: 10.1007/978-1-4939-0709-0_12. Methods Mol Biol. 2014. PMID: 24788269 Review.
Cited by
-
InCoB2013 introduces Systems Biology as a major conference theme.BMC Syst Biol. 2013 Nov 4;7 Suppl 3(Suppl 3):S1. doi: 10.1186/1752-0509-7-S3-S1. BMC Syst Biol. 2013. PMID: 24555777 Free PMC article.
-
Ontology-supported research on vaccine efficacy, safety and integrative biological networks.Expert Rev Vaccines. 2014 Jul;13(7):825-41. doi: 10.1586/14760584.2014.923762. Epub 2014 Jun 7. Expert Rev Vaccines. 2014. PMID: 24909153 Free PMC article. Review.
-
ENQUIRE automatically reconstructs, expands, and drives enrichment analysis of gene and Mesh co-occurrence networks from context-specific biomedical literature.PLoS Comput Biol. 2025 Feb 11;21(2):e1012745. doi: 10.1371/journal.pcbi.1012745. eCollection 2025 Feb. PLoS Comput Biol. 2025. PMID: 39932993 Free PMC article.
-
Associating disease-related genetic variants in intergenic regions to the genes they impact.PeerJ. 2014 Oct 23;2:e639. doi: 10.7717/peerj.639. eCollection 2014. PeerJ. 2014. PMID: 25374782 Free PMC article.
-
HNdb: an integrated database of gene and protein information on head and neck squamous cell carcinoma.Database (Oxford). 2016 Mar 24;2016:baw026. doi: 10.1093/database/baw026. Print 2016. Database (Oxford). 2016. PMID: 27013077 Free PMC article.
References
-
- The PubMed database. http://www.ncbi.nlm.nih.gov/pubmed/
-
- MeSH fact sheet. URL: http://www.nlm.nih.gov/pubs/factsheets/mesh.html, accessed on March 23, 2013.
-
- Blaschke C, Andrade MA, Ouzounis C, Valencia A. Automatic extraction of biological information from scientific text: protein-protein interactions. Proc Int Conf Intell Syst Mol Biol. 1999. pp. 60–67. - PubMed
-
- Jenssen TK, Laegreid A, Komorowski J, Hovig E. A literature network of human genes for high-throughput analysis of gene expression. Nat Genet. 2001;7(1):21–28. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

