Quartet-based methods to reconstruct phylogenetic networks
- PMID: 24555518
- PMCID: PMC3941989
- DOI: 10.1186/1752-0509-8-21
Quartet-based methods to reconstruct phylogenetic networks
Abstract
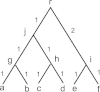
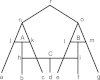
Background: Phylogenetic networks are employed to visualize evolutionary relationships among a group of nucleotide sequences, genes or species when reticulate events like hybridization, recombination, reassortant and horizontal gene transfer are believed to be involved. In comparison to traditional distance-based methods, quartet-based methods consider more information in the reconstruction process and thus have the potential to be more accurate.
Results: We introduce QuartetSuite, which includes a set of new quartet-based methods, namely QuartetS, QuartetA, and QuartetM, to reconstruct phylogenetic networks from nucleotide sequences. We tested their performances and compared them with other popular methods on two simulated nucleotide sequence data sets: one generated from a tree topology and the other from a complicated evolutionary history containing three reticulate events. We further validated these methods to two real data sets: a bacterial data set consisting of seven concatenated genes of 36 bacterial species and an influenza data set related to recently emerging H7N9 low pathogenic avian influenza viruses in China.
Conclusion: QuartetS, QuartetA, and QuartetM have the potential to accurately reconstruct evolutionary scenarios from simple branching trees to complicated networks containing many reticulate events. These methods could provide insights into the understanding of complicated biological evolutionary processes such as bacterial taxonomy and reassortant of influenza viruses.
Figures





References
-
- Nelson M, Vibound C, Simonsen L, Bennett R, Griesemer S, George K, Taylor J, Spiro D, Sengamalay NA, Ghedin E, Taubenberger J, Holmes E. Multiple reassortment events in the evolutionary history of h1n1 influenza a virus since 1918. PLOS Pathog. 2008;4(2):1000012. doi: 10.1371/journal.ppat.1000012. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

