Annotating N termini for the human proteome project: N termini and Nα-acetylation status differentiate stable cleaved protein species from degradation remnants in the human erythrocyte proteome
- PMID: 24555563
- PMCID: PMC3979129
- DOI: 10.1021/pr401191w
Annotating N termini for the human proteome project: N termini and Nα-acetylation status differentiate stable cleaved protein species from degradation remnants in the human erythrocyte proteome
Abstract
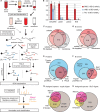
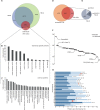
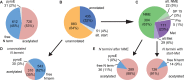
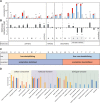
A goal of the Chromosome-centric Human Proteome Project is to identify all human protein species. With 3844 proteins annotated as "missing", this is challenging. Moreover, proteolytic processing generates new protein species with characteristic neo-N termini that are frequently accompanied by altered half-lives, function, interactions, and location. Enucleated and largely void of internal membranes and organelles, erythrocytes are simple yet proteomically challenging cells due to the high hemoglobin content and wide dynamic range of protein concentrations that impedes protein identification. Using the N-terminomics procedure TAILS, we identified 1369 human erythrocyte natural and neo-N-termini and 1234 proteins. Multiple semitryptic N-terminal peptides exhibited improved mass spectrometric identification properties versus the intact tryptic peptide enabling identification of 281 novel erythrocyte proteins and six missing proteins identified for the first time in the human proteome. With an improved bioinformatics workflow, we developed a new classification system and the Terminus Cluster Score. Thereby we described a new stabilizing N-end rule for processed protein termini, which discriminates novel protein species from degradation remnants, and identified protein domain hot spots susceptible to cleavage. Strikingly, 68% of the N-termini were within genome-encoded protein sequences, revealing alternative translation initiation sites, pervasive endoproteolytic processing, and stabilization of protein fragments in vivo. The mass spectrometry proteomics data have been deposited to ProteomeXchange with the data set identifier <PXD000434>.
Figures

References
-
- Paik Y.-K.; Jeong S.-K.; Omenn G. S.; Uhlen M.; Hanash S.; Cho S. Y.; Lee H.-J.; Na K.; Choi E.-Y.; Yan F.; Zhang F.; Zhang Y.; Snyder M.; Cheng Y.; Chen R.; Marko-Varga G.; Deutsch E. W.; Kim H.; Kwon J.-Y.; Aebersold R.; Bairoch A.; Taylor A. D.; Kim K. Y.; Lee E.-Y.; Hochstrasser D.; Legrain P.; Hancock W. S. The Chromosome-Centric Human Proteome Project for cataloging proteins encoded in the genome. Nat. Biotechnol. 2012, 30, 221–223. - PubMed
-
- Kleifeld O.; Doucet A.; Auf dem Keller U.; Prudova A.; Schilling O.; Kainthan R. K.; Starr A. E.; Foster L. J.; Kizhakkedathu J. N.; Overall C. M. Isotopic labeling of terminal amines in complex samples identifies protein N-termini and protease cleavage products. Nat. Biotechnol. 2010, 28, 281–288. - PubMed
-
- Wilson J. G.; Wong W. W.; Murphy E. E. 3.; Schur P. H.; Fearon D. T. Deficiency of the C3b/C4b receptor (CR1) of erythrocytes in systemic lupus erythematosus: analysis of the stability of the defect and of a restriction fragment length polymorphism of the CR1 gene. J. Immunol. 1987, 138, 2708–2710. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

