Groupwise multi-atlas segmentation of the spinal cord's internal structure
- PMID: 24556080
- PMCID: PMC4009677
- DOI: 10.1016/j.media.2014.01.003
Groupwise multi-atlas segmentation of the spinal cord's internal structure
Abstract
The spinal cord is an essential and vulnerable component of the central nervous system. Differentiating and localizing the spinal cord internal structure (i.e., gray matter vs. white matter) is critical for assessment of therapeutic impacts and determining prognosis of relevant conditions. Fortunately, new magnetic resonance imaging (MRI) sequences enable clinical study of the in vivo spinal cord's internal structure. Yet, low contrast-to-noise ratio, artifacts, and imaging distortions have limited the applicability of tissue segmentation techniques pioneered elsewhere in the central nervous system. Additionally, due to the inter-subject variability exhibited on cervical MRI, typical deformable volumetric registrations perform poorly, limiting the applicability of a typical multi-atlas segmentation framework. Thus, to date, no automated algorithms have been presented for the spinal cord's internal structure. Herein, we present a novel slice-based groupwise registration framework for robustly segmenting cervical spinal cord MRI. Specifically, we provide a method for (1) pre-aligning the slice-based atlases into a groupwise-consistent space, (2) constructing a model of spinal cord variability, (3) projecting the target slice into the low-dimensional space using a model-specific registration cost function, and (4) estimating robust segmentation susing geodesically appropriate atlas information. Moreover, the proposed framework provides a natural mechanism for performing atlas selection and initializing the free model parameters in an informed manner. In a cross-validation experiment using 67 MR volumes of the cervical spinal cord, we demonstrate sub-millimetric accuracy, significant quantitative and qualitative improvement over comparable multi-atlas frameworks, and provide insight into the sensitivity of the associated model parameters.
Keywords: Cervical spinal cord segmentation; Groupwise registration; Multi-atlas segmentation; Spinal cord internal structure.
Copyright © 2014 Elsevier B.V. All rights reserved.
Figures


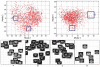


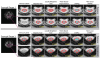
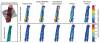

References
-
- Aljabar P, Heckemann R, Hammers A, Hajnal J, Rueckert D. Multi-atlas based segmentation of brain images: Atlas selection and its effect on accuracy. Neuroimage. 2009;46:726–738. - PubMed
-
- Amari S, Cichocki A, Yang HH. A new learning algorithm for blind signal separation. Advances in neural information processing systems. 1996:757–763.
-
- Artaechevarria X, Muñoz-Barrutia A, Ortiz-de-Solorzano C. Combination strategies in multi-atlas image segmentation: Application to brain MR data. IEEE Trans. Med. Imaging. 2009;28:1266–1277. - PubMed
Publication types
MeSH terms
Grants and funding
- 1R21NS064534/NS/NINDS NIH HHS/United States
- 2R01EB006136/EB/NIBIB NIH HHS/United States
- R21 NS064534/NS/NINDS NIH HHS/United States
- T32 EB014841/EB/NIBIB NIH HHS/United States
- ImNIH/Intramural NIH HHS/United States
- K01 EB009120/EB/NIBIB NIH HHS/United States
- R01 EB006193/EB/NIBIB NIH HHS/United States
- 1R03EB012461/EB/NIBIB NIH HHS/United States
- 1T32EB014841/EB/NIBIB NIH HHS/United States
- R01 EB006136/EB/NIBIB NIH HHS/United States
- R03 EB012461/EB/NIBIB NIH HHS/United States
- R01EB006193/EB/NIBIB NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

