Preparing an annotated gold standard corpus to share with extramural investigators for de-identification research
- PMID: 24556292
- PMCID: PMC4125487
- DOI: 10.1016/j.jbi.2014.01.014
Preparing an annotated gold standard corpus to share with extramural investigators for de-identification research
Abstract
Objective: The current study aims to fill the gap in available healthcare de-identification resources by creating a new sharable dataset with realistic Protected Health Information (PHI) without reducing the value of the data for de-identification research. By releasing the annotated gold standard corpus with Data Use Agreement we would like to encourage other Computational Linguists to experiment with our data and develop new machine learning models for de-identification. This paper describes: (1) the modifications required by the Institutional Review Board before sharing the de-identification gold standard corpus; (2) our efforts to keep the PHI as realistic as possible; (3) and the tests to show the effectiveness of these efforts in preserving the value of the modified data set for machine learning model development.
Materials and methods: In a previous study we built an original de-identification gold standard corpus annotated with true Protected Health Information (PHI) from 3503 randomly selected clinical notes for the 22 most frequent clinical note types of our institution. In the current study we modified the original gold standard corpus to make it suitable for external sharing by replacing HIPAA-specified PHI with newly generated realistic PHI. Finally, we evaluated the research value of this new dataset by comparing the performance of an existing published in-house de-identification system, when trained on the new de-identification gold standard corpus, with the performance of the same system, when trained on the original corpus. We assessed the potential benefits of using the new de-identification gold standard corpus to identify PHI in the i2b2 and PhysioNet datasets that were released by other groups for de-identification research. We also measured the effectiveness of the i2b2 and PhysioNet de-identification gold standard corpora in identifying PHI in our original clinical notes.
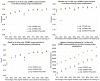
Results: Performance of the de-identification system using the new gold standard corpus as a training set was very close to training on the original corpus (92.56 vs. 93.48 overall F-measures). Best i2b2/PhysioNet/CCHMC cross-training performances were obtained when training on the new shared CCHMC gold standard corpus, although performances were still lower than corpus-specific trainings.
Discussion and conclusion: We successfully modified a de-identification dataset for external sharing while preserving the de-identification research value of the modified gold standard corpus with limited drop in machine learning de-identification performance.
Keywords: Automated de-identification; De-identification gold standard; Health insurance portability and accountability act; Natural Language Processing; Privacy of patient data; Protected Health Information.
Copyright © 2014 The Authors. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Gupta D, Saul M, Gilbertson J. Evaluation of a deidentification (De-Id) software engine to share pathology reports and clinical documents for research. American journal of clinical pathology. 2004;121(2):176–86. Epub 2004/02/27. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

