Bacteria that glide with helical tracks
- PMID: 24556443
- PMCID: PMC3964879
- DOI: 10.1016/j.cub.2013.12.034
Bacteria that glide with helical tracks
Abstract
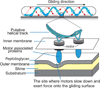
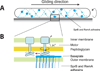
Many bacteria glide smoothly on surfaces, despite having no discernable propulsive organelles on their surface. Recent experiments with Myxococcus xanthus and Flavobacterium johnsoniae show that both of these distantly related bacterial species glide using proteins that move in helical tracks, albeit with significantly different motility mechanisms. Both species utilize proton-motive force for movement. Although the motors that power gliding in M. xanthus have been identified, the F. johnsoniae motors remain to be discovered.
Copyright © 2014 Elsevier Ltd. All rights reserved.
Figures
References
-
- Berg HC. The rotary motor of bacterial flagella. Annu Rev Biochem. 2003;72:19–54. - PubMed
-
- Lauga E. Life around the scallop theorem. Soft Matter. 2011;7
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

