Blockade of BFA-mediated apoptosis in macrophages by the HIV-1 Nef protein
- PMID: 24556695
- PMCID: PMC3944234
- DOI: 10.1038/cddis.2014.16
Blockade of BFA-mediated apoptosis in macrophages by the HIV-1 Nef protein
Expression of concern in
-
Editorial Expression of Concern to: Blockade of BFA-mediated apoptosis in macrophages by the HIV-1 Nef protein.Cell Death Dis. 2025 Apr 7;16(1):260. doi: 10.1038/s41419-025-07600-5. Cell Death Dis. 2025. PMID: 40195299 Free PMC article. No abstract available.
Abstract
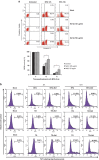
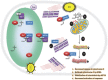
HIV-1 Nef protein has key roles at almost all stages of the viral life cycle. We assessed the role of Nef and of the translation elongation factor eEF1A in primary human macrophages. Nuclear retention experiments and inhibition of the exportin-t (Exp-t) pathway suggested that cytoplasmic relocalization of eEF1A, mediated by Exp-t occurs in Nef-treated monocyte-derived macrophages (MDMs). We observed the presence of tRNA in the Nef/eEF1A complexes. Nucleocytoplasmic relocalization of the Nef/eEF1A complexes prevented stress-induced apoptosis of MDMs treated with brefeldin A. Blockade of stress-induced apoptosis of MDMs treated with HIV-1 Nef resulted from enhanced nucleocytoplasmic transport of eEF1A with decreased release of mitochondrial cytochrome c, and from increased tRNA binding to cytochrome c, ultimately leading to an inhibition of caspase activation. Our results indicate that HIV-1 Nef, through the nucleocytoplasmic relocalization of eEF1A and tRNAs, enhances resistance to stress-induced apoptosis in primary human macrophages.
Figures







References
-
- Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. - PubMed
-
- Masutani H, Ueda S, Yodoi J. The thioredoxin system in retroviral infection and apoptosis. Cell Death Diff. 2005;12:991–998. - PubMed
-
- Uhm HD, Orenstein JM, Wahl SM. Fas mediates apoptosis in human monocytes by a reactive oxygen intermediate dependent pathway. J Immunol. 1996;156:3469–3477. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

