An unbiased proteomic screen reveals caspase cleavage is positively and negatively regulated by substrate phosphorylation
- PMID: 24556848
- PMCID: PMC4014278
- DOI: 10.1074/mcp.M113.037374
An unbiased proteomic screen reveals caspase cleavage is positively and negatively regulated by substrate phosphorylation
Abstract
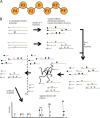
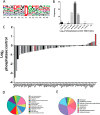
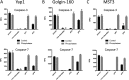
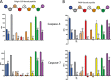
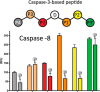
Post-translational modifications of proteins regulate diverse cellular functions, with mounting evidence suggesting that hierarchical cross-talk between distinct modifications may fine-tune cellular responses. For example, in apoptosis, caspases promote cell death via cleavage of key structural and enzymatic proteins that in some instances is inhibited by phosphorylation near the scissile bond. In this study, we systematically investigated how protein phosphorylation affects susceptibility to caspase cleavage using an N-terminomic strategy, namely, a modified terminal amino isotopic labeling of substrates (TAILS) workflow, to identify proteins for which caspase-catalyzed cleavage is modulated by phosphatase treatment. We validated the effects of phosphorylation on three of the identified proteins and found that Yap1 and Golgin-160 exhibit decreased cleavage when phosphorylated, whereas cleavage of MST3 was promoted by phosphorylation. Furthermore, using synthetic peptides we systematically examined the influence of phosphoserine throughout the entirety of caspase-3, -7, and -8 recognition motifs and observed a general inhibitory effect of phosphorylation even at residues considered outside the classical consensus motif. Overall, our work demonstrates a role for phosphorylation in controlling caspase-mediated cleavage and shows that N-terminomic strategies can be tailored to study cross-talk between phosphorylation and proteolysis.
Figures

References
-
- Hanahan D., Weinberg R. A. (2000) The hallmarks of cancer. Cell 100, 57–70 - PubMed
-
- Hunter T. (2000) Signaling—2000 and beyond. Cell 100, 113–127 - PubMed
-
- Taylor R. C., Cullen S. P., Martin S. J. (2008) Apoptosis: controlled demolition at the cellular level. Nat. Rev. Mol. Cell. Biol. 9, 231–241 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

