Homology-directed repair of DNA nicks via pathways distinct from canonical double-strand break repair
- PMID: 24556991
- PMCID: PMC3956201
- DOI: 10.1073/pnas.1400236111
Homology-directed repair of DNA nicks via pathways distinct from canonical double-strand break repair
Abstract
DNA nicks are the most common form of DNA damage, and if unrepaired can give rise to genomic instability. In human cells, nicks are efficiently repaired via the single-strand break repair pathway, but relatively little is known about the fate of nicks not processed by that pathway. Here we show that homology-directed repair (HDR) at nicks occurs via a mechanism distinct from HDR at double-strand breaks (DSBs). HDR at nicks, but not DSBs, is associated with transcription and is eightfold more efficient at a nick on the transcribed strand than at a nick on the nontranscribed strand. HDR at nicks can proceed by a pathway dependent upon canonical HDR factors RAD51 and BRCA2; or by an efficient alternative pathway that uses either ssDNA or nicked dsDNA donors and that is strongly inhibited by RAD51 and BRCA2. Nicks generated by either I-AniI or the CRISPR/Cas9(D10A) nickase are repaired by the alternative HDR pathway with little accompanying mutagenic end-joining, so this pathway may be usefully applied to genome engineering. These results suggest that alternative HDR at nicks may be stimulated in physiological contexts in which canonical RAD51/BRCA2-dependent HDR is compromised or down-regulated, which occurs frequently in tumors.
Keywords: cancer; gene conversion; loss of heterozygosity; recombination; targeted gene correction.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

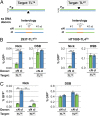

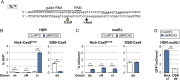
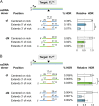
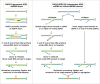
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

