Reticulon 4 is necessary for endoplasmic reticulum tubulation, STIM1-Orai1 coupling, and store-operated calcium entry
- PMID: 24558039
- PMCID: PMC3969502
- DOI: 10.1074/jbc.M114.548602
Reticulon 4 is necessary for endoplasmic reticulum tubulation, STIM1-Orai1 coupling, and store-operated calcium entry
Abstract
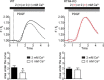
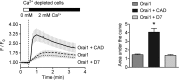
Despite recent advances in understanding store-operated calcium entry (SOCE) regulation, the fundamental question of how ER morphology affects this process remains unanswered. Here we show that the loss of RTN4, is sufficient to alter ER morphology and severely compromise SOCE. Mechanistically, we show this to be the result of defective STIM1-Orai1 coupling because of loss of ER tubulation and redistribution of STIM1 to ER sheets. As a functional consequence, RTN4-depleted cells fail to sustain elevated cytoplasmic Ca(2+) levels via SOCE and therefor are less susceptible to Ca(2+) overload induced apoptosis. Thus, for the first time, our results show a direct correlation between ER morphology and SOCE and highlight the importance of RTN4 in cellular Ca(2+) homeostasis.
Keywords: Apoptosis; Cell Signaling; Endoplasmic Reticulum (ER); Imaging; Membrane; reticulons.
Figures










References
-
- Lee C., Chen L. B. (1988) Dynamic behavior of endoplasmic reticulum in living cells. Cell 54, 37–46 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

