Probing the unknowns in cytokinin-mediated immune defense in Arabidopsis with systems biology approaches
- PMID: 24558299
- PMCID: PMC3929428
- DOI: 10.4137/BBI.S13462.
Probing the unknowns in cytokinin-mediated immune defense in Arabidopsis with systems biology approaches
Abstract
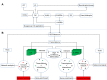
Plant hormones involving salicylic acid (SA), jasmonic acid (JA), ethylene (Et), and auxin, gibberellins, and abscisic acid (ABA) are known to regulate host immune responses. However, plant hormone cytokinin has the potential to modulate defense signaling including SA and JA. It promotes plant pathogen and herbivore resistance; underlying mechanisms are still unknown. Using systems biology approaches, we unravel hub points of immune interaction mediated by cytokinin signaling in Arabidopsis. High-confidence Arabidopsis protein-protein interactions (PPI) are coupled to changes in cytokinin-mediated gene expression. Nodes of the cellular interactome that are enriched in immune functions also reconstitute sub-networks. Topological analyses and their specific immunological relevance lead to the identification of functional hubs in cellular interactome. We discuss our identified immune hubs in light of an emerging model of cytokinin-mediated immune defense against pathogen infection in plants.
Keywords: cytokinin; gene expression; interaction networks; plant hormones; systems biology.
Figures



Similar articles
-
Signaling Crosstalk between Salicylic Acid and Ethylene/Jasmonate in Plant Defense: Do We Understand What They Are Whispering?Int J Mol Sci. 2019 Feb 4;20(3):671. doi: 10.3390/ijms20030671. Int J Mol Sci. 2019. PMID: 30720746 Free PMC article. Review.
-
Two-component elements mediate interactions between cytokinin and salicylic acid in plant immunity.PLoS Genet. 2012 Jan;8(1):e1002448. doi: 10.1371/journal.pgen.1002448. Epub 2012 Jan 26. PLoS Genet. 2012. PMID: 22291601 Free PMC article.
-
The impact of cytokinin on jasmonate-salicylate antagonism in Arabidopsis immunity against infection with Pst DC3000.Plant Signal Behav. 2013 Oct;8(10):doi: 10.4161/psb.26791. doi: 10.4161/psb.26791. Plant Signal Behav. 2013. PMID: 24494231 Free PMC article.
-
Melatonin and its relationship to plant hormones.Ann Bot. 2018 Feb 12;121(2):195-207. doi: 10.1093/aob/mcx114. Ann Bot. 2018. PMID: 29069281 Free PMC article. Review.
-
Ethylene and jasmonic acid signaling affect the NPR1-independent expression of defense genes without impacting resistance to Pseudomonas syringae and Peronospora parasitica in the Arabidopsis ssi1 mutant.Mol Plant Microbe Interact. 2003 Jul;16(7):588-99. doi: 10.1094/MPMI.2003.16.7.588. Mol Plant Microbe Interact. 2003. PMID: 12848424
Cited by
-
Intervention of Phytohormone Pathways by Pathogen Effectors.Plant Cell. 2014 Jun;26(6):2285-2309. doi: 10.1105/tpc.114.125419. Epub 2014 Jun 10. Plant Cell. 2014. PMID: 24920334 Free PMC article. Review.
-
Cytokinin Production by Azospirillum brasilense Contributes to Increase in Growth, Yield, Antioxidant, and Physiological Systems of Wheat (Triticum aestivum L.).Front Microbiol. 2022 May 19;13:886041. doi: 10.3389/fmicb.2022.886041. eCollection 2022. Front Microbiol. 2022. PMID: 35663903 Free PMC article.
-
Large Scale Proteomic Data and Network-Based Systems Biology Approaches to Explore the Plant World.Proteomes. 2018 Jun 3;6(2):27. doi: 10.3390/proteomes6020027. Proteomes. 2018. PMID: 29865292 Free PMC article.
-
Cytokinin Signaling Downstream of the His-Asp Phosphorelay Network: Cytokinin-Regulated Genes and Their Functions.Front Plant Sci. 2020 Nov 17;11:604489. doi: 10.3389/fpls.2020.604489. eCollection 2020. Front Plant Sci. 2020. PMID: 33329676 Free PMC article. Review.
-
A combined tissue-engineered/in silico signature tool patient stratification in lung cancer.Mol Oncol. 2018 Aug;12(8):1264-1285. doi: 10.1002/1878-0261.12323. Epub 2018 Jun 22. Mol Oncol. 2018. PMID: 29797762 Free PMC article.
References
-
- Hwang I, Sheen J, Muller B. Cytokinin signaling networks. Annu Rev Plant Biol. 2012;63:353–80. - PubMed
-
- Argueso CT, Raines T, Kieber JJ. Cytokinin signaling and transcriptional networks. Curr Opin Plant Biol. 2010;13(5):533–9. - PubMed
-
- Wilkinson S, Kudoyarova GR, Veselov DS, Arkhipova TN, Davies WJ. Plant hormone interactions: innovative targets for crop breeding and management. J Exp Bot. 2012;63(9):3499–509. - PubMed
LinkOut - more resources
Full Text Sources

