Model of a DNA-protein complex of the architectural monomeric protein MC1 from Euryarchaea
- PMID: 24558431
- PMCID: PMC3928310
- DOI: 10.1371/journal.pone.0088809
Model of a DNA-protein complex of the architectural monomeric protein MC1 from Euryarchaea
Abstract
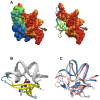
In Archaea the two major modes of DNA packaging are wrapping by histone proteins or bending by architectural non-histone proteins. To supplement our knowledge about the binding mode of the different DNA-bending proteins observed across the three domains of life, we present here the first model of a complex in which the monomeric Methanogen Chromosomal protein 1 (MC1) from Euryarchaea binds to the concave side of a strongly bent DNA. In laboratory growth conditions MC1 is the most abundant architectural protein present in Methanosarcina thermophila CHTI55. Like most proteins that strongly bend DNA, MC1 is known to bind in the minor groove. Interaction areas for MC1 and DNA were mapped by Nuclear Magnetic Resonance (NMR) data. The polarity of protein binding was determined using paramagnetic probes attached to the DNA. The first structural model of the DNA-MC1 complex we propose here was obtained by two complementary docking approaches and is in good agreement with the experimental data previously provided by electron microscopy and biochemistry. Residues essential to DNA-binding and -bending were highlighted and confirmed by site-directed mutagenesis. It was found that the Arg25 side-chain was essential to neutralize the negative charge of two phosphates that come very close in response to a dramatic curvature of the DNA.
Conflict of interest statement
Figures






Similar articles
-
New protein-DNA complexes in archaea: a small monomeric protein induces a sharp V-turn DNA structure.Sci Rep. 2019 Oct 3;9(1):14253. doi: 10.1038/s41598-019-50211-2. Sci Rep. 2019. PMID: 31582767 Free PMC article.
-
Chemical shifts assignments of the archaeal MC1 protein and a strongly bent 15 base pairs DNA duplex in complex.Biomol NMR Assign. 2015 Apr;9(1):215-7. doi: 10.1007/s12104-014-9577-8. Epub 2014 Sep 12. Biomol NMR Assign. 2015. PMID: 25212183
-
Atypical recognition of particular DNA sequences by the archaeal chromosomal MC1 protein.Biochemistry. 2005 Aug 2;44(30):10369-77. doi: 10.1021/bi0474416. Biochemistry. 2005. PMID: 16042414
-
Generalized electrostatic model of the wrapping of DNA around oppositely charged proteins.Biopolymers. 2007 Jun 5;86(2):127-35. doi: 10.1002/bip.20711. Biopolymers. 2007. PMID: 17330872 Review.
-
Nonsequence-specific DNA recognition: a structural perspective.Structure. 2000 Apr 15;8(4):R83-9. doi: 10.1016/s0969-2126(00)00126-x. Structure. 2000. PMID: 10801483 Review.
Cited by
-
New protein-DNA complexes in archaea: a small monomeric protein induces a sharp V-turn DNA structure.Sci Rep. 2019 Oct 3;9(1):14253. doi: 10.1038/s41598-019-50211-2. Sci Rep. 2019. PMID: 31582767 Free PMC article.
-
Genome-wide transcriptional response to silver stress in extremely halophilic archaeon Haloferax alexandrinus DSM 27206 T.BMC Microbiol. 2023 Dec 4;23(1):381. doi: 10.1186/s12866-023-03133-z. BMC Microbiol. 2023. PMID: 38049746 Free PMC article.
-
BioSpring: An elastic network framework for interactive exploration of macromolecular mechanics.Protein Sci. 2025 May;34(5):e70130. doi: 10.1002/pro.70130. Protein Sci. 2025. PMID: 40249023 Free PMC article.
-
DNA-Binding Properties of African Swine Fever Virus pA104R, a Histone-Like Protein Involved in Viral Replication and Transcription.J Virol. 2017 May 26;91(12):e02498-16. doi: 10.1128/JVI.02498-16. Print 2017 Jun 15. J Virol. 2017. PMID: 28381576 Free PMC article.
-
The Hypersaline Archaeal Histones HpyA and HstA Are DNA Binding Proteins That Defy Categorization According to Commonly Used Functional Criteria.mBio. 2023 Apr 25;14(2):e0344922. doi: 10.1128/mbio.03449-22. Epub 2023 Feb 13. mBio. 2023. PMID: 36779711 Free PMC article.
References
-
- Luijsterburg MS, White MF, van Driel R, Dame RT (2008) The major architects of chromatin: architectural proteins in bacteria, archaea and eukaryotes. Crit Rev Biochem Mol Biol 43: 393–418. - PubMed
-
- Reeve JN (2003) Archaeal chromatin and transcription. Mol Microbiol 48: 587–598. - PubMed
-
- White MF, Bell SD (2002) Holding it together: chromatin in the Archaea. Trends Genet 18: 621–626. - PubMed
-
- Sandman K, Reeve JN (2005) Archaeal chromatin proteins: different structures but common function? Curr Opin Microbiol 8: 656–661. - PubMed
-
- DeLange RJ, Green GR, Searcy DG (1981) A histone-like protein (HTa) from Thermoplasma acidophilum. I. Purification and properties. J Biol Chem 256: 900–904. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

