FKBP12.6 activates RyR1: investigating the amino acid residues critical for channel modulation
- PMID: 24559985
- PMCID: PMC3945099
- DOI: 10.1016/j.bpj.2013.12.041
FKBP12.6 activates RyR1: investigating the amino acid residues critical for channel modulation
Abstract
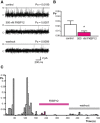
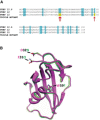
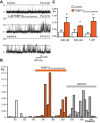
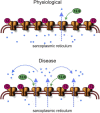
We have previously shown that FKBP12 associates with RyR2 in cardiac muscle and that it modulates RyR2 function differently to FKBP12.6. We now investigate how these proteins affect the single-channel behavior of RyR1 derived from rabbit skeletal muscle. Our results show that FKBP12.6 activates and FKBP12 inhibits RyR1. It is likely that both proteins compete for the same binding sites on RyR1 because channels that are preactivated by FKBP12.6 cannot be subsequently inhibited by FKBP12. We produced a mutant FKBP12 molecule (FKBP12E31Q/D32N/W59F) where the residues Glu(31), Asp(32), and Trp(59) were converted to the corresponding residues in FKBP12.6. With respect to the functional regulation of RyR1 and RyR2, the FKBP12E31Q/D32N/W59F mutant lost all ability to behave like FKBP12 and instead behaved like FKBP12.6. FKBP12E31Q/D32N/W59F activated RyR1 but was not capable of activating RyR2. In conclusion, FKBP12.6 activates RyR1, whereas FKBP12 activates RyR2 and this selective activator phenotype is determined within the amino acid residues Glu(31), Asp(32), and Trp(59) in FKBP12 and Gln(31), Asn(32), and Phe(59) in FKBP12.6. The opposing but different effects of FKBP12 and FKBP12.6 on RyR1 and RyR2 channel gating provide scope for diversity of regulation in different tissues.
Copyright © 2014 The Authors. Published by Elsevier Inc. All rights reserved.
Figures




References
-
- Marx S.O., Reiken S., Marks A.R. PKA phosphorylation dissociates FKBP12.6 from the calcium release channel (ryanodine receptor): defective regulation in failing hearts. Cell. 2000;101:365–376. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

