Effect of C-terminal protein tags on pentitol and L-arabinose transport by Ambrosiozyma monospora Lat1 and Lat2 transporters in Saccharomyces cerevisiae
- PMID: 24561586
- PMCID: PMC3993308
- DOI: 10.1128/AEM.04067-13
Effect of C-terminal protein tags on pentitol and L-arabinose transport by Ambrosiozyma monospora Lat1 and Lat2 transporters in Saccharomyces cerevisiae
Abstract
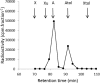
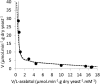
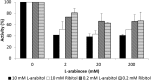
Functional expression in heterologous hosts is often less successful for integral membrane proteins than for soluble proteins. Here, two Ambrosiozyma monospora transporters were successfully expressed in Saccharomyces cerevisiae as tagged proteins. Growth of A. monospora on l-arabinose instead of glucose caused transport activities of l-arabinose, l-arabitol, and ribitol, measured using l-[1-(3)H]arabinose, l-[(14)C]arabitol, and [(14)C]ribitol of demonstrated purity. A. monospora LAT1 and LAT2 genes were cloned earlier by using their ability to improve the growth of genetically engineered Saccharomyces cerevisiae on l-arabinose. However, the l-arabinose and pentitol transport activities of S. cerevisiae carrying LAT1 or LAT2 are only slightly greater than those of control strains. S. cerevisiae carrying the LAT1 or LAT2 gene fused in frame to the genes for green fluorescent protein (GFP) or red fluorescent protein (mCherry) or adenylate kinase (AK) exhibited large (>3-fold for LAT1; >20-fold for LAT2) increases in transport activities. Lat1-mCherry transported l-arabinose with high affinity (Km ≈ 0.03 mM) and l-arabitol and ribitol with very low affinity (Km ≥ 75 mM). The Lat2-GFP, Lat2-mCherry, and Lat2-AK fusion proteins could not transport l-arabinose but were high-affinity pentitol transporters (Kms ≈ 0.2 mM). The l-arabinose and pentitol transport activities of A. monospora could not be completely explained by any combination of the observed properties of tagged Lat1 and Lat2, suggesting either that tagging and expression in a foreign membrane alters the transport kinetics of Lat1 and/or Lat2 or that A. monospora contains at least one more l-arabinose transporter.
Figures


Similar articles
-
Cloning of two genes (LAT1,2) encoding specific L: -arabinose transporters of the L: -arabinose fermenting yeast Ambrosiozyma monospora.Appl Biochem Biotechnol. 2011 Jul;164(5):604-11. doi: 10.1007/s12010-011-9161-y. Epub 2011 Jan 21. Appl Biochem Biotechnol. 2011. PMID: 21253888
-
Novel transporters from Kluyveromyces marxianus and Pichia guilliermondii expressed in Saccharomyces cerevisiae enable growth on L-arabinose and D-xylose.Yeast. 2015 Oct;32(10):615-28. doi: 10.1002/yea.3084. Epub 2015 Aug 17. Yeast. 2015. PMID: 26129747
-
Functional Analysis of Two l-Arabinose Transporters from Filamentous Fungi Reveals Promising Characteristics for Improved Pentose Utilization in Saccharomyces cerevisiae.Appl Environ Microbiol. 2015 Jun 15;81(12):4062-70. doi: 10.1128/AEM.00165-15. Epub 2015 Apr 3. Appl Environ Microbiol. 2015. PMID: 25841015 Free PMC article.
-
Metabolic Engineering for Improved Fermentation of L-Arabinose.J Microbiol Biotechnol. 2019 Mar 28;29(3):339-346. doi: 10.4014/jmb.1812.12015. J Microbiol Biotechnol. 2019. PMID: 30786700 Review.
-
[Progress in research of pentose transporters and C6/C5 co-metabolic strains in Saccharomyces cerevisiae].Sheng Wu Gong Cheng Xue Bao. 2018 Oct 25;34(10):1543-1555. doi: 10.13345/j.cjb.180031. Sheng Wu Gong Cheng Xue Bao. 2018. PMID: 30394022 Review. Chinese.
Cited by
-
Current strategies for protein production and purification enabling membrane protein structural biology.Biochem Cell Biol. 2016 Dec;94(6):507-527. doi: 10.1139/bcb-2015-0143. Epub 2016 Jan 20. Biochem Cell Biol. 2016. PMID: 27010607 Free PMC article. Review.
-
Engineering of Pentose Transport in Saccharomyces cerevisiae for Biotechnological Applications.Front Bioeng Biotechnol. 2020 Jan 29;7:464. doi: 10.3389/fbioe.2019.00464. eCollection 2019. Front Bioeng Biotechnol. 2020. PMID: 32064252 Free PMC article. Review.
-
The Penicillium chrysogenum transporter PcAraT enables high-affinity, glucose-insensitive l-arabinose transport in Saccharomyces cerevisiae.Biotechnol Biofuels. 2018 Mar 13;11:63. doi: 10.1186/s13068-018-1047-6. eCollection 2018. Biotechnol Biofuels. 2018. PMID: 29563966 Free PMC article.
-
Membrane Protein Production and Purification from Escherichia coli and Sf9 Insect Cells.Methods Mol Biol. 2020;2168:3-49. doi: 10.1007/978-1-0716-0724-4_1. Methods Mol Biol. 2020. PMID: 33582985
-
In vitro resynthesis of lichenization reveals the genetic background of symbiosis-specific fungal-algal interaction in Usnea hakonensis.BMC Genomics. 2020 Sep 29;21(1):671. doi: 10.1186/s12864-020-07086-9. BMC Genomics. 2020. PMID: 32993496 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

